1. Background
Viral hepatitis is one of the most significant public health problems, caused mainly by hepatitis viruses A through E (1-4). Amongst hepatitis viruses, Hepatitis B virus (HBV) can cause lifelong infection and it was estimated that 350 million people are chronically infected with HBV around the world. It is endemic in Asia and most people in the region have been infected with HBV during their childhood (5). Recently, Iran has moved from intermediate rate of HBV in the past to the current low HBV seroprevalence of 1.7%, and still one of the main national health priorities is decreasing HBV frequency in the society (6-8). HBV infected individuals are prone to the development of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (9-12). However, the exact mechanisms of persistent HBV infection have not been fully elucidated, nevertheless defects in host cellular immune responses are crucial in causing viral persistence (13).
Production of cytokines is regulated by molecular mechanisms that have been attributed to variations in genetic components at the transcriptional, posttranscriptional, and translational levels (14). MicroRNAs are evolutionary conserved, single-stranded non-coding RNA molecules of about 22 to 25 nucleotides (15). These small RNAs are well known for their posttranscriptional gene regulation, provided that microRNAs bind to their target site at 3’UTR region of protein-coding mRNAs. This binding can result in mRNA cleavage, inhibition of translation, or stimulate poly (A) tail removal, which leads to reduced levels of mRNA concentration (16, 17). The IL-16 is one of the pro-inflammatory cytokines (18) located on chromosome 15.q26.3, and expresses a precursor protein composed of 631 amino acids, which is then cleaved by caspase-3 to form the functional C-terminal domain with 121 amino acids (19). It can activate T cells, monocytes, macrophages, and dendritic cells by binding to the main receptors (CD4 molecule) presented on these cells (20, 21). Furthermore, it can stimulate the expression of other proinflammatory cytokines, including IL-1β, IL-6, and IL-15 (22). In the recent years, the association of IL-16 polymorphisms with different diseases such as prostate, gastric, colon cancer (23, 24), and Crohn’s disease were investigated (20); likewise in one study the association of IL-16 polymorphisms and risk of HBV-related hepatocellular carcinoma was evaluated (25). However, to our knowledge, the relationship between this cytokine polymorphism and chronic HBV infection susceptibility has not been reported to date.
2. Objectives
Thus, the aim of this study was to investigate the association of rs1131445 variation, in the MicroRNA binding site of IL-16 located at the 3’UTR of the gene, in patients with chronic HBV infection and in control subjects.
3. Patients and Methods
3.1. Study Population
A total of 531 subjects, including 262 chronic HBV patients confirmed as HBV positive, and HCV, HDV and human immunodeficiency virus negative and 269 healthy controls, were entered in this case-control study. The enrolled subjects were individuals referred to the Taleghani hospital of Tehran from January 2010 to October 2011. The cases and controls sample size was calculated based on a previously published formula (26, 27). Informed consent was obtained from all subjects at the time of enrollment and the ethical review boards of the institution approved the study protocol.
3.2. Serological Assays
Routine biochemical tests were performed using automated techniques. HBsAg, anti-HBs, and anti-HBc were detected by the enzyme linked immunosorbent assay (ELISA) technique (Diapro, Italy). Antibodies to hepatitis C virus, hepatitis D virus, and human immunodeficiency virus were detected by routine ELISA technique (Diapro, Italy).
3.3. Genotyping
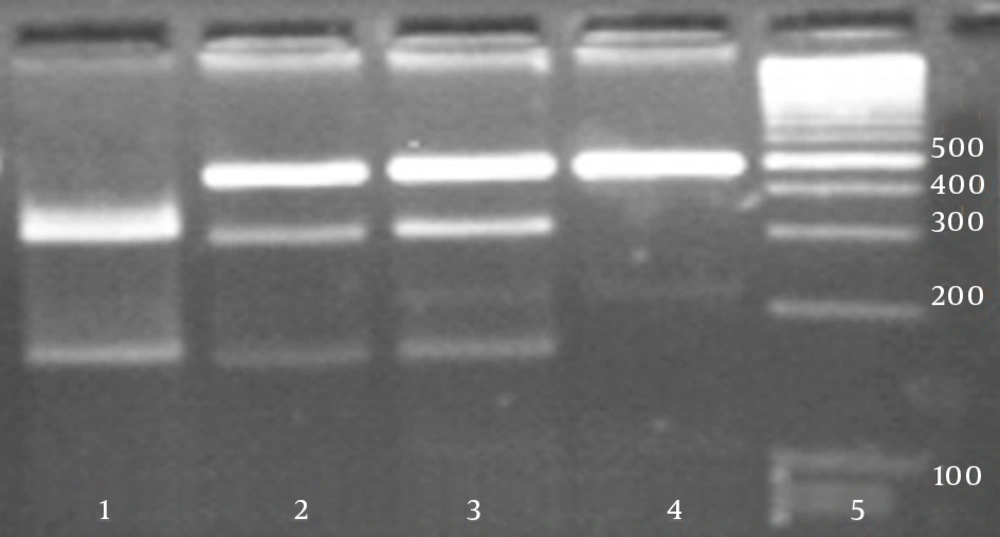
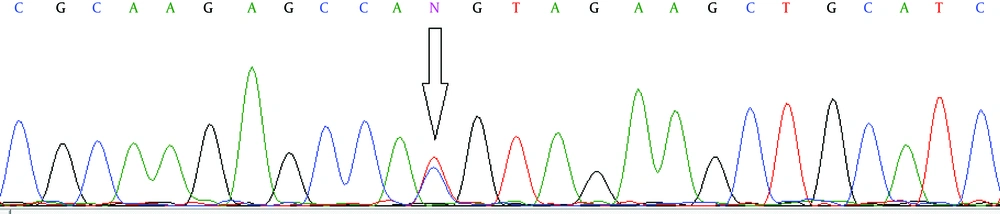
Total DNA was isolated from whole blood, using a standard protocol for extraction with phenol-chloroform. For each studied gene, polymorphic regions were amplified by the polymerase chain reaction (PCR) in a total volume of 25 μL, using PCR reagents (the annealing temperature for the PCR was 65°C), followed by digestion of the PCR product with a corresponding restriction enzyme (RFLP). Primer sets (Bioneer, South Korea) and restriction enzyme BsaAI (New England Biolabs, England) used for the PCR-RFLP assay are shown in Table 1. Product and allele sizes are also given in Table 1. Fifteen microliters of the PCR product was then cleaved with a corresponding restriction enzyme. The PCR and restriction enzyme products were analyzed separately by electrophoresis on 2% and 3% agarose (Roche, Germany) gel, respectively and subsequently stained with ethidium bromide to be visualized under exposure of UV light. Additionally, to confirm the PCR-RFLP assay, about 10% of total samples for each genotype were randomly selected and then their PCR products were sequenced on an ABI genetic analyzer 3130xl (Applied Biosystems, USA).
| Gene Name | Primers Sequence (5’ → 3’) | SNP | Position | PCR Product Size, bp | Restriction Enzyme | RFLP Fragments Size, bp |
|---|---|---|---|---|---|---|
| IL-16 | rs1131445 | 3’UTR | 460 | BsaAI | Genotype TT: 460, Genotype CT: 460 + 300 + 160, Genotype CC: 300 + 160 | |
| Forward | GAGATCATTCACTCATACATCTGG | |||||
| Reverse | TCATATACACGCTGGTTCCTTCTG |
3.4. Statistical Methods
The association between patients and controls for genotype and allele frequencies was determined by the χ2 test. To adjust for confounding factors including age and gender, logistic regression analysis was used. Odds ratios (OR) and 95% Confidence Intervals (95% CI) were used to estimate the association between individual polymorphisms and chronic HBV. P-values of less than 0.05 were considered statistically significant. All statistical analyses were performed using the SPSS software (version 15.0; SPSS, Chicago, IL, USA).
4. Results
In this research the case group consisted of 262 chronic HBV patients, including 164 male and 98 female individuals, and the control group consisted of 269 healthy individuals with 155 males and 114 females. The results of the RFLP assay are given in Figure 1. No significant difference was found between males and females in neither group (Table 2). The genotype and allelic frequencies of IL-16 gene variation of the patient and control groups are presented in Table 3. The frequencies of genotypes in the case group were 44.3%, 48.1%, and 7.6% for TT, TC and CC genotypes, respectively, while in the control group these frequencies were 51.3%, 38.7%, and 10%. No statistically significant differences were shown in the distributions of genotypes and allele frequencies between the patient and control groups for rs1131445 polymorphism (P = 0.084). However, after adjustment for covariates including age and gender, a significant difference was found in the TC genotype between the cases and controls (OR: 0.696, CI: 0.485 - 0.997, P = 0.048). All sequencing results were rechecked by the sequencing method and were concordant with the original RFLP results (Figure 2).
| Variables | Controls (n = 269) | Cases (n = 262) | P Value |
|---|---|---|---|
| Age | 46.06 ± 17.390 | 44.34 ± 15.898 | 0.233 |
| Gender | 0.242 | ||
| Male | 155 (57.6) | 164 (62.6) | |
| Female | 114 (42.4) | 98 (37.4) |
aData are presented as mean ± SD or No. (%).
| IL-16 SNP (rs1131445) | Controls (n= 269) | Cases (n = 262) | OR ( 95% CI) | P Value |
|---|---|---|---|---|
| Genotypes | ||||
| TT | 138 (51.3) | 116 (44.3) | 1.00 (reference) | |
| TC | 104 (38.7) | 126 (48.1) | 0.696 (0.485 - 0.997) | 0.048 |
| CC | 27 (10) | 20 (7.6) | 1.148 (0.608 - 2.171) | 0.670 |
| Alleles | ||||
| T | 380 (70.6) | 358 (68.3) | 1.00 (reference) | |
| C | 158 (29.4) | 166 (31.7) | 0.911 (0.701 - 1.184) | 0.487 |
aAdjusted for gender and age.
bData are presented as No. (%).
5. Discussion
Cytokines play a critical role in immune and inflammatory responses. However, cytokine coding genes are polymorphic to a great extent, which means some of these polymorphisms can affect the expression of cytokines. Single nucleotide polymorphisms (SNPs) are the most frequent types of genetic variations. In this regard, SNPs within cytokine genes can affect the severity and progression of immune-mediated and chronic inflammatory diseases (28). Furthermore, SNPs in microRNA-binding sites can alter microRNA-mRNA interaction that results in an increase or decrease in protein translation (17). To the best of our knowledge, there are no previous studies about the association of rs1131445 in the microRNA-binding site of the IL-16 gene and susceptibility to chronic HBV infection.
Dendritic cells, as a group of antigen-presenting cells, can build a bridge between pathogens and the T-cell system. This mechanism also has been described for viral diseases such as Hepatitis B infection (29, 30). On the other hand, IL-16 activates dendritic cells by binding to CD4 molecules (21). It is also well understood that CD8+ T-cells have an important role in controlling HBV replication (31), at the same time, activation of CD8+ T-cells can be carried out by IL-16 (21). T helper 1 and T helper 2 cells are two distinct T helper cell subsets, which produce cytokines to regulate the immune system. The IL-16 belongs to T helper 1 regulatory cytokines with the ability of expanding inflammation in T helper1-mediated hypersensitivity (32, 33). T helper 1 cell products such as IL-2, activate cytotoxic T lymphocytes to enhance viral clearance. This can also be seen in HBV infection (34, 35). The host immune responses to hepatitis viruses in order to clear the viruses result in T helper 1-mediated inflammatory response, while IL-16 enhances this response. Additionally, during HBV infection, IL-16 may participate in killing virus-infected cells and minimizing cellular damage (32, 33).
In this study, the results indicated that the TC genotype of the rs1131445 could significantly increase the risk of HBV chronic infection. In only one study conducted by Li et al. it was demonstrated that TG and GG genotypes of rs11556218 were associated with a significantly decreased risk of chronic hepatitis B compared with the TT genotype (25). The association between IL-16 gene polymorphism and several diseases have been investigated previously (25, 36-38). Gu et al. reported that SNPs and haplotypes of the IL-16 gene including rs1131445 were significantly associated with susceptibility to Graves’ disease (39). Similar studies on different study populations may result in different outcomes. Landi et al. found no significant association between rs1131445 polymorphism of IL-16 gene and colorectal cancer in their study population from the Czech Republic (16), while Azimzadeh et al. reported a significant association between this polymorphism and colorectal cancer in their study amongst an Iranian population (40).
Overall, our study may involve a number of possible limitations. The first one is the small sample size that prevents drawing strong conclusions. The second one is the existence of other polymorphisms outside our studied region that have an influence on the function of the studied polymorphism. Therefore, results may be misleading when other interacting variations are not considered. In summary, the results of this investigation clarified that the presence of the IL-16 TC genotype was associated with chronic HBV infection. These results should be replicated in other investigations with an appropriate number of subjects to further evaluate the potential association between this polymorphism and HBV susceptibility.