1. Background
Tuberculosis (TB) is an old disease that has been known as one of the major causes of morbidity and mortality worldwide (1, 2). Regardless, one of every three humans is infected with the TB pathogen. According to the world health organization (WHO), 7.1 billion people worldwide are infected with TB and more than 20 million people suffer from active TB. Annually, more than 8 million people are infected with TB and about 3 million TB patients lose their lives (3, 4). Therefore, new and effective vaccines against adult pulmonary TB are a global requirement, and research in this field has expanded widely. Although the bacillus calmette guerin (BCG) vaccine offers protection against TB in childhood (5), and may protect against disseminated TB or TB meningitis, it still has some limitations, including reduction in protection over time, low level of protection against pulmonary TB, side effects such as lymphadenitis and disseminated life-threatening infection if administrated to immunocompromised patients (6).
Most studies have focused on live attenuated vaccines and protein subunit vaccines. In many cases, DNA vaccines containing Mycobacterium tuberculosis antigens have been used to induce protection against primary infection with the pathogen. Further, these vaccines have also been used as boosters after BCG vaccination (7). Antigens such as: heat shock protein (HSP) 60, Hsp70, Ag85, ESAT-6, and CFP10 are new TB vaccine candidates as well as new diagnostic factors (8). The epitopes encoded by DNA vaccines are expressed on the cells through MHC molecules. Helper and killer T lymphocytes recognize these antigen-MHCI complexes. Therefore, DNA vaccines induce strong CD4+ (Th1) and (CTL) CD8+ responses against M. tuberculosis (9). DNA vaccines are used for the prevention and treatment of infectious diseases and cancer. Selection of highly immunogenic and conserved antigens is a critical step in the preparation of such vaccines.
Culture-filtered proteins (CFPs) are important in the prevention and diagnosis of TB (10). Genomic analysis has shown that CFP10 contains 99 amino acids, and has a molecular weight of 10.7 kDa (11). The C-terminal region of CFP10 can attach to macrophages and monocytes, and is susceptible to digestion with trypsin (12). CFP10 or the CFP10/ESAT6 complex induces the release of TNFα from J774 macrophages and Th1 cells. TNFα is essential to invoke the immune cells to eliminate the pathogens in granuloma. It should be noted that depending of its level, TNFα can either induce host cell death or prevent it (13).
2. Objectives
The purpose of this study was to construct a vector containing cfp10 of M. tuberculosis strain H37Rv.
3. Materials and Methods
3.1. DNA Extraction
Mycobacterium tuberculosis strain H37Rv was cultured in 7H9. Stock cultures were stored at -70˚C. Some colonies were also cultured on Lowenstein Jensen medium and were incubated at 37˚C. After 14 days, 20 colonies were isolated and homogenized in 400 µL of buffer (pH 7.5) containing Tris (Merck, Germany) (100 mM) and Tween (Merck, Germany) 20 (0.05%). Then, 20 µL of proteinase k (18.5 mg/mL, Fermentas, Germany) was added and the mixture was incubated for 3 hour at 55˚C. For enzyme inactivation and DNA extraction, the samples were boiled for 10 minutes, followed by centrifugation at 14,000 rpm for 10 minutes. Next, the supernatant containing the DNA was stored until it was used for amplification.
3.2. Polymerase Chain Reaction for Amplification cfp10
The primers were designed using the software Gene Runner. The PCR reaction mixture contained the following ingredients: 10 pmol forward primer (5′-ATTGATAGAATTCGCAGAGATGAAGACCGATGCCGCT-3′) and reverse primer (5′-TCATTAATCTAGATTATCAGAAGCCCATTTGCGAGGACAG-3′) (Cinnagen, Iran) for cfp10, 1 μL of DNA sample, 1.5 mM MgCl2, 0.2 mM each dNTP, and 5 U/μL Taq polymerase. The final reaction volume was 25 μL. The genomic DNA of M. tuberculosis strain H37Rv was used as a template at a concentration of 100 ng/μL. The first cycle of the PCR reaction was performed for 5 minutes at 95°C, followed by 35 cycles of 1 minute at 95°C, 1 minute at 65°C, and 1 at minute 72°C. The final cycle was performed for 7 minutes at 72°C. Subsequently, 50 μL of the PCR product was electrophoresed on 1% agarose gel in Tris/borate/EDTA buffer (pH 8). To purify the cfp10 fragment, approximately 150 mg of the gel was cut and purified using the Invisorb spin DNA extraction kit (Invisorb, Germany) according to the manufacturer’s instructions.
3.3. Cfp10 Cloning in Vector pcDNA3.1 (+)
PCR product and pcDNA3.1 (+) plasmid were digested with EcoRI and XbaI (Fermentas, Germany). These enzymes have restriction site each in pcDNA 3.1 (+). The reaction was performed using 20 μL of cfp10 DNA (25 ng/μL), 5 μL of Buffer H, 2 μL of EcoRI (10 U/μL), 2 μL of XbaI (10 U/μL), and 31 μL of deuterium-depleted water (DDW). The selection of the best buffer for performance of any restriction enzyme is important. The vector concentration was 100 ng/μL after enzyme digestion. The digested products were purified from the gel using the gel extraction kit. In order to insert cfp10 into pcDNA3.1 (+) vector, ligation was performed using T4 DNA ligase (14). The ligation reaction mixture contained: 6 μL of pcDNA3.1(+), 12 μL of cfp10, 2.5 μL of T4 DNA ligase buffer, 2 μL of T4 DNA ligase (5 U/μL) Fermentas, Germany, 2 μL of the polyethylene glycol Fermentas, Germany, and 0.5 μL of DDW.
Escherichia coli strain JM109 treated with the cold 0.1 M CaCl2/MgCl2 solution for acquiring DNA(15). This strain was selected because it is appropriate for cloning. Calcium deposits the gene on the cell wall of the bacterium. Competent cells were transformed using the heat shock method provided by Sambrook et al. (16). The transformed bacteria were cultured on LB agar containing ampicillin 100 μg/mL. Colony-PCR was performed to confirm colonies containing the recombinant vector. Next, the recombinant vector was extracted using a QIAprep Miniprep Kit (Qiagen, USA). The cloning accuracy was confirmed using enzyme digestion with EcoRI (10 U/μL) followed by sequencing (DNAMAN software). Enzymatic digestion was performed using 10 μL of recombinant vector (100 ng/μL), 5 μL of Buffer H, 2 μL of EcoRI (10 U/μL), and 33 μL of DDW.
4. Results
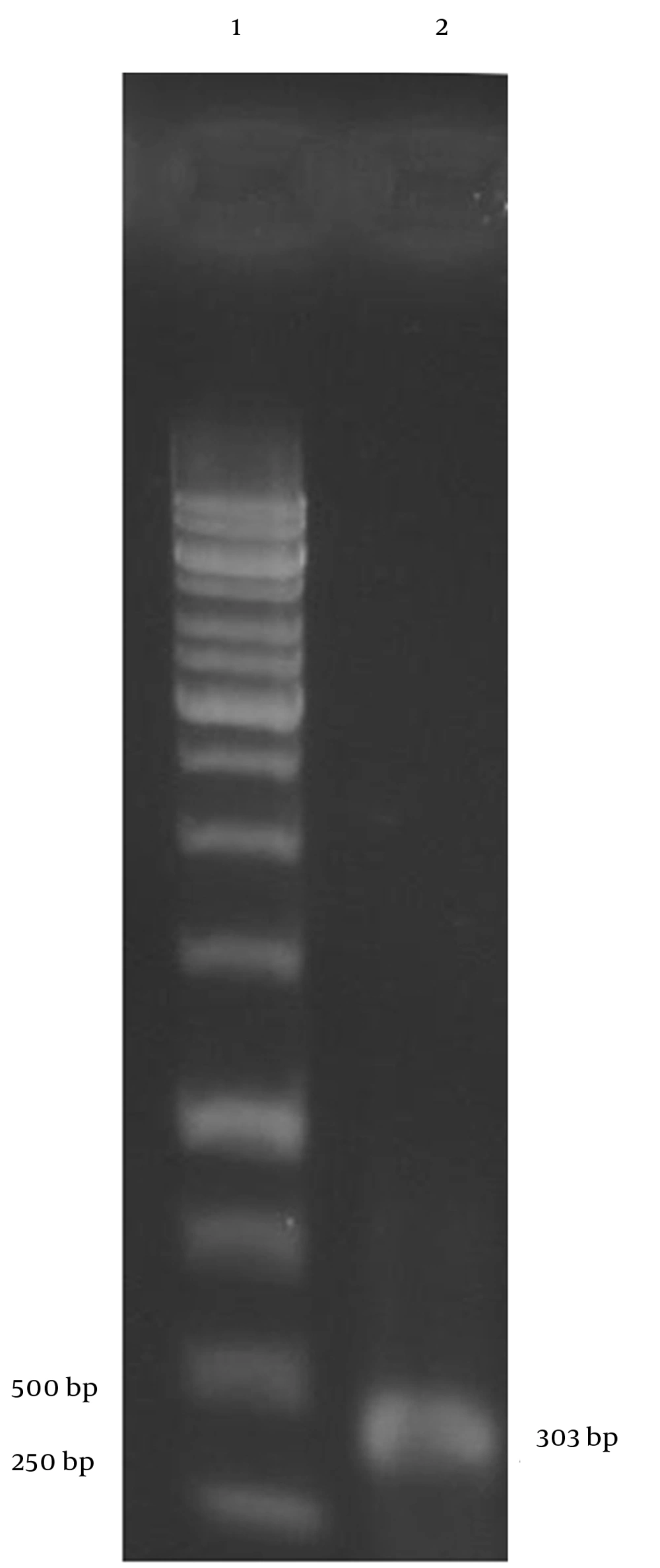
We amplified cfp10M. tuberculosis strain H37Rv using PCR. A 303-bp fragment was obtained following 1% w/v agarose gel electrophoresis (Figure 1).
The PCR product was electrophoresed after digestion with EcoRI and XbaI. Digestion with each enzyme was performed using the specific buffer. A fragment without any considerable size change was observed. The pcDNA3.1 (+) vector was also digested with the same enzymes, and a 5428-bp band was observed. In the next step, the target gene was inserted into the backbone using T4 DNA ligase. The circular product yielded a band with a relative molecular weight on the gel. The ligation product was used to transform E. coli strain JM109 with cold 0.1 M CaCl2/MgCl2 solution, and colony-PCR was performed using the forward and reverse primers for cfp10 to confirm gene insertion into pcDNA3.1 (+). The PCR product was gel electrophoresed and a fragment of 303 bp was observed. The recombinant vector was purified using the QIAprep Miniprep Kit and subjected to digestion with EcoRI. A band of 5730 bp was observed upon digestion. To confirm cfp10 insertion into the vector, simultaneous digestion with EcoRI and XbaI was also carried out, and two fragments were observed: one band (5428 bp) was identical to linearized vector and the other band was consistent with cfp10 (303 bp). Successful cloning was finally confirmed by sequencing the recombinant vector.
5. Discussion
To date, BCG has been used as a vaccine against TB. However, its effect on pulmonary TB is debatable. Therefore, new effective, safe, and more reliable vaccines with preferably new modes of action are needed. DNA vaccines using antigens of M. tuberculosis are candidates for future vaccines (17). A study has shown that DNA vaccines provide protection against M. bovis in animal models. However, this protection is found only when mycobacterial DNA is coupled with adjuvants or DNA encoding co-stimulatory molecules such as CD80 and CD86. The immunity induced by this vaccine is not equivalent to the protection offered by BCG. Of note, if this vaccine is administered as a booster after BCG, then it would elicit a more effective immune response than that achieved using BCG vaccination alone (18). Because TB is one of the most dangerous infectious diseases, there is an urgent need for a better vaccine than BCG. Furthermore, in order to control this disease, it is necessary to design a stronger vaccine than BCG and/or a vaccine that is capable of boosting the immunogenicity of or the immune response elicited by BCG.
The advantages of direct immunization with plasmid DNA encoding antigens of M. tuberculosis include sustainability, easy preparation and handling, and safety for immune-compromised patients. In addition, such vaccines could be stored at room temperature and could be administrated repeatedly to boost immunity (19). Mammalian expression vectors can be injected directly into muscle cells, and as a result of continuous transcription and translation of the genes, a strong immune response is can be elicited (20).
CFPs of M. tuberculosis secreted during bacterial growth phase are the major targets of the T cells. In addition, Th1 cytokines and TNFα are major immune mediators against M. tuberculosis in mice and humans. These cytokines are essential for the expression of inducible nitric oxide synthase, which is involved in the immune response against infection in mice. Culture filtered antigens of M. tuberculosis stimulate the immune system to varying degrees. These antigens have been shown to induce a protective immune response in a model of BALB/c mice, especially in the late phase of bacterial infection, which might be related to the high density of the antigen in this phase (21).
Mahairas and colleagues demonstrated for the first time in 1996 the existence of specific genomic regions in M. tuberculosis. They studied genetic differences between M. tuberculosis, M. bovis, and BCG using genomic subtractive hybridization. Their results showed that 3 specific genomic regions that are present in M. tuberculosis and M. bovis are absent in BCG. These deleted regions are called regions of difference or regions deleted or briefly RDs (RD1, RD2, and RD3) (22). Evidences suggest that the protein encoded by RD1 is recognized by the immune system and has particular importance in the immune response against TB due to its strong antigenicity (23, 24).
This region harbors cfp10 in the genome of M. tuberculosis. It is noted that Cfp10 is always in complex with Esat6. Cloning and sequence analysis of cfp10 was carried out in 1989. Further analyses revealed that the encoded protein contains 99 amino acids and a molecular weight of 10.7 kDa. The epitopes of the product were shown to be associated with T cells (11). In one study, plasmids pcDNA3.1 (+)/esat6 and pcDNA3.1 (+)/cfp10 were constructed and injected into BALB/c mice and the RNA expression in mouse cells was verified by RT-PCR. Further investigations revealed that these vectors could induce proliferation of lymphocytes in the vaccinated mice (25).
In our study, the vector pcDNA3.1 (+)/cfp10 was designed and cloning was confirmed using a prokaryote system. Due to the fact that it is a eukaryotic shuttle vector, RNA expression pattern in a eukaryotic system should be investigated in further research. DNA vaccine studies generally focus on Ag85a and ESAT6. However, in the present study, we used the antigen cfp10 for designing the recombinant vector. In attempts to design a protective DNA vaccine some other antigens have also been evaluated. For example, in a study conducted by Nabavinia et al. (26) Mtb72f was subcloned into pET21b vector, and E. coli BL21 (DE3) was used to express the protein. Technically, in their study, 4 enzymes were employed to join the 3 genes, and finally protein expression was analyzed using western blot in a prokaryotic system. In contrast, in our study, EcoRI and XbaI were used for cloning the gene into pcDNA3.1 (+) vector. Cloning accuracy was confirmed by colony-PCR, enzyme digestion, and sequencing. In colony-PCR, the size of the cloned fragment with cfp10 primers was found to be accurate. Restriction enzyme digestion showed that the fragment separated from the vector was cfp10. Finally, DNA sequencing with cfp10 primers confirmed the cloning. We propose to expand this study by expression of CFP10 protein and investigation of its immunogenicity in mouse.
To summarize, here we cloned cfp10 into a eukaryotic expression system for use as a vaccine. In future, studies could be carried out in order to purify the CFP10 protein and subsequent monitoring of the production of IFN-γ, TNFα, and IgG1 against CFP10 in animal models, which might lead to promising findings for human administration, although most DNA vaccines of bacteria have not reached the clinical phase.