1. Background
The accumulation of fibrin clot in blood vessels induces thrombosis, which leads to cardiovascular diseases. Thrombosis is an abnormal blood clot inside a blood vessel. The blood clots can detach from the vascular wall and travel in the blood. These free floating thrombi (now called emboli) can lodge anywhere in the cardiovascular system, including the lungs or brain (as in a thrombotic stroke). Fibrin is implicated in many heart attacks, since cardiac arrest usually occurs after a plaque’s cap fractures, causing a blood clot to form over the fracture and block blood flow.
Thrombolytic therapy has become a conventional treatment for myocardial infarction (AMI), yet at present, clinically prescribed thrombolytic drugs have problems such as delayed action and other side effects like bleeding, re occlusion etc. (1). A variety of fibrinolytic enzymes, such as tissue plasminogen activator (t-PA), urokinase and streptokinase have been extensively used as thrombolytic agents (2). Several investigations are presently being pursued to enhance the efficacy and specificity of fibrinolytic therapy. Fibrinolytic enzymes have attracted interest as thrombolytic agents because of their efficiency in the fibrinolytic process including plasmin activation. Nattokinase (NK) is a potent fibrinolytic agent for thrombosis therapy, which was purified from natto, a traditional Japanese fermented food, which not only directly lyses thrombi in vivo, but can also enhance fibrinolytic activity in plasma and increase the amount of t-PA by oral administration (3-6). Previous studies have shown the production of NK from Bacillus spp. and this species is the most involved in enzyme production. Despite this, only one study has reported NK production from Pseudomonas sp. isolated from soil (6). Hence this study is unique and the findings are the first report on the production of NK from p. aeruginosa CMSS isolated from cow milk.
2. Objectives
The main aim of this study was to enhance the production of NK from P. aeruginosa CMSS using optimization and a strain improvement method.
3. Materials and Methods
3.1. Isolation and Screening of Nattokinase-Producing Pseudomonas Species
A total of 15 cow milk samples were collected from the Vellore district of Tamil Nadu, India and the samples were serially diluted to 10 -7. The spread plating technique was performed using 100 µL of 10 - 5and 10 - 6 dilutions on 5% blood agar medium, (Hi-Media, India) and incubated at 37˚C for 24 hours. β-hemolytic colonies were selected based on colony morphology and Grams reaction. The isolated organisms were screened for NK production by casein hydrolytic assay. Skimmed milk agar (Hi-Media, India) plate was supplemented with 200 µL of plasma. The isolated Pseudomonas sp. was streaked on the skim milk agar medium, and incubated at 37˚C for 12 hours (7, 8).
3.2. Strain Selection and Identification
The potent bacterial strain (CMSS) isolated from cow milk was characterized morphologically and biochemically according to Bergey’s manual of determinative bacteriology (9). Pure cultures of the identified strain was maintained on nutrient agar plates and stored at 4˚C.
3.3. Molecular Characterization of the CMSS Strain
The significant step in the analysis of phylogenetic relationship between species is DNA sequence comparison (10). The individual colonies of the potent isolate (CMSS) were picked up with a sterilized tooth pick, suspended in 0.5 µL of sterile saline in a 1.5 mL centrifuge tube and centrifuged at 10,000 rpm for ten minutes. The pellet was suspended in 0.5 µL of Insta Gene Matrix (Bio-Rad, USA) and incubated at 56˚C for 30 minutes and then heated at 100˚C for 10 minutes. After heating, the supernatant was used for the polymerase chain reaction. The amplification of the DNA was done using universal forward and reverse primers, 27F (5’- AGAGTTTGATCMTGGCTCAG -3’) and 1492R (5’- TACGGYTACCTTGTTACGACTT- 3’), respectively.
The conditions for amplification were 35 cycles at 94˚C for 45 seconds, 55˚C for 60 seconds, and 72˚C for 60 seconds (11). The PCR products were sequenced using forward and reverse primers 518F (5’- CCAGCAGCCGCGGTAATACG -3’) and 800R (5’- TACCAGGGTATCTAATCC -3’), respectively. Sequencing was performed using the Big Dye terminator cycle sequencing kit (Applied Bio Systems, USA). Sequencing products were resolved on an Applied Bio systems model 3730XL automated DNA sequencing system (Applied Bio Systems, USA). The 16S rRNA sequences were integrated into the database with the automatic alignment tool. A phylogenetic tree was generated by performing distance matrix analysis, using the neighbor-joining method (12). Database search and comparisons were done with a BLAST search using the National Center for Biotechnology Information (NCBI) database (13).
3.4. Production of Nattokinase
The CMSS isolate isolated from cow milk was used for the NK production. The 1% seed inoculums were inoculated in to three different medium containing 100 mL of nutrient broth, production medium (1% shrimp shell, 0.1% KH2PO4, 0.05% MgSO4, pH-7 at 37˚C) and nutrient broth (Hi-Media) with 0.1% shrimp shell and incubated at 37˚C for 24 hours in a shaker at 120 rpm. The medium was then centrifuged at 10,000 rpm for 10 minutes and the supernatant was used as the crude enzyme extract. The enzyme from the three different media was analyzed by radial caseinolytic plate assay (7, 8).
3.5. Optimization of Conditions for Enhanced Nattokinase Production
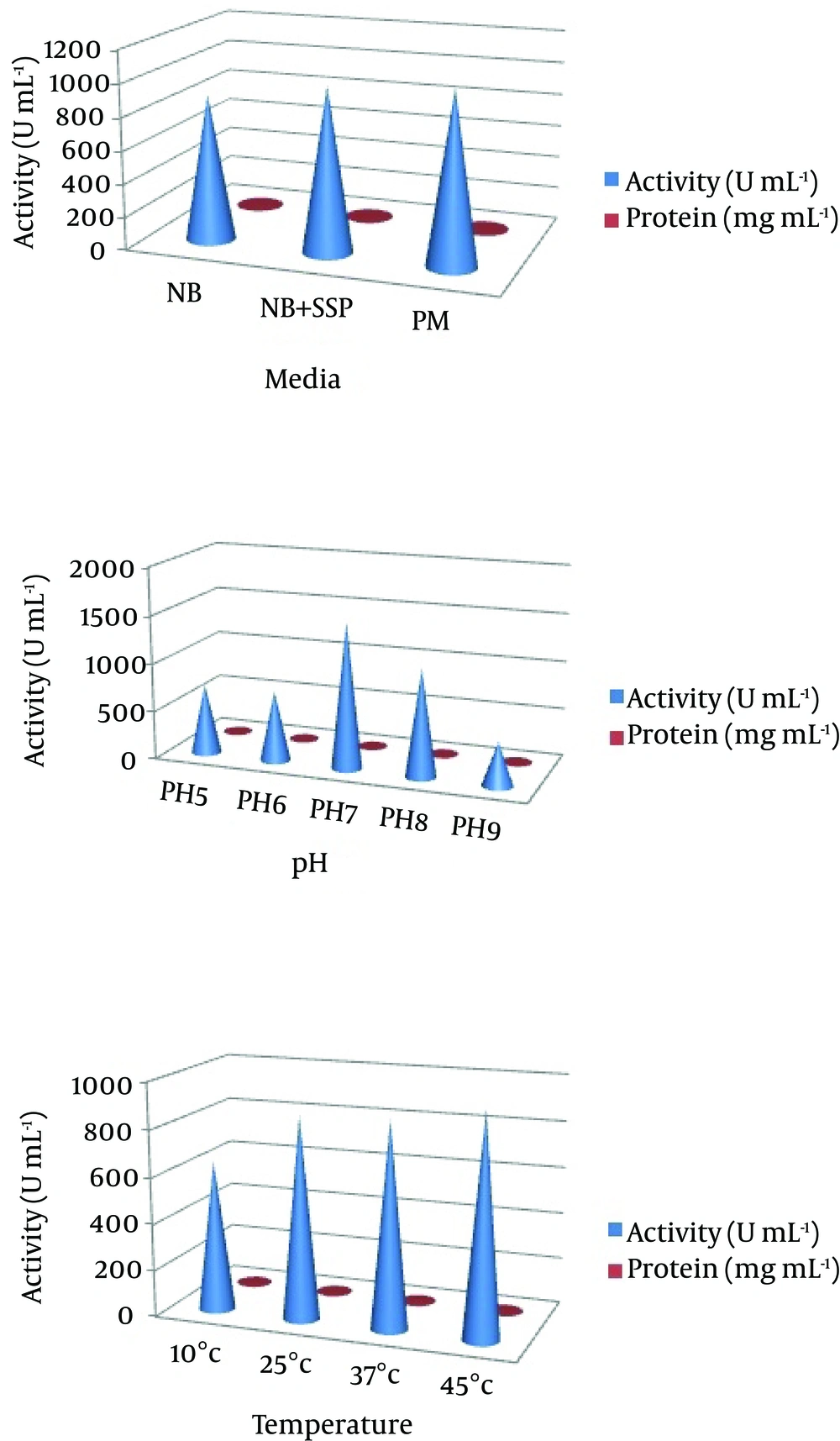
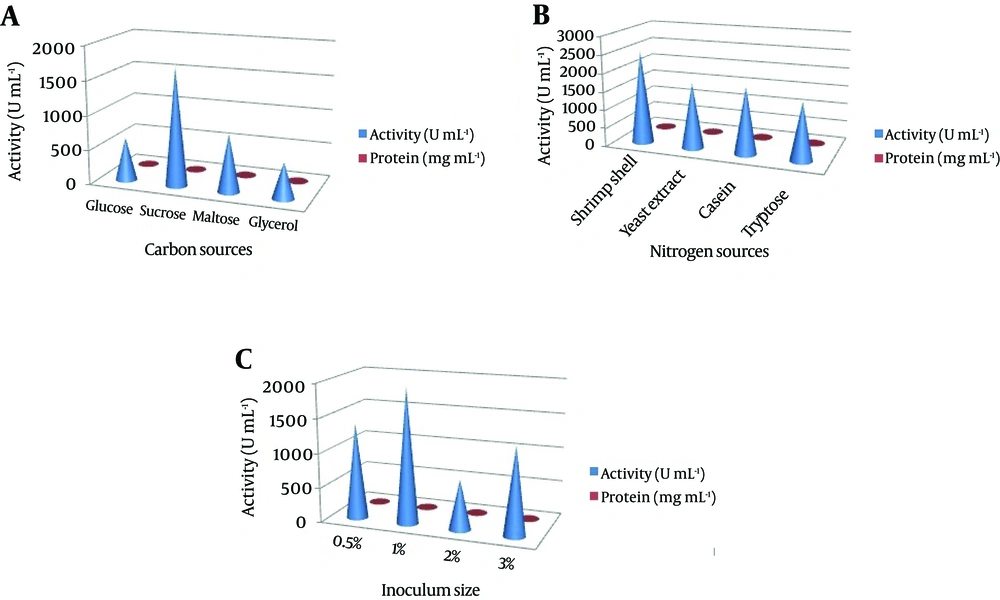
The production of NK depends on several process parameters. These parameters vary for microbial strains being employed for the production, and hence conditions such as pH, temperature, carbon, nitrogen, and inoculums size have been optimized to ensure maximum enzyme production. The effect of various carbon sources such as glucose, sucrose, maltose and glycerol at 1% concentration were assessed in the production medium. Various nitrogen sources such as shrimp shell powder, yeast extract, casein and tryptone at the 1% concentration were used. The effect of temperature was studied at 10˚C, 25˚C, 37˚C and 45˚C. Similarly, the effect of pH on NK production was investigated by varying the pH of the medium to pH 5.0, 6.0, 7.0, 8.0 and 9.0. Various concentrations of inoculum size with respect to NK production were tested by adopting different concentrations of 0.5%, 1%, 2% and 3%. The culture medium in a 250 mL Erlenmeyer flask was aerobically incubated at 37˚C for 24 hours on a rotary shaker (120 rpm). At the end of cultivation, the crude enzyme was obtained by centrifugation at 10,000 rpm and 4˚C for 20 minutes.
3.6. Strain Improvement by Random UV-Mutagenesis
Strain improvement was done by random UV-mutagenesis. The wild-type strain was prepared in phosphate buffer (pH 7.0) and 4 mL inoculum was pipetted aseptically into sterile petri dishes of 80 mm diameter having a flat bottom. Exposure was carried out at a distance of 20 cm away from the center of the germicidal lamp (UV light source) with occasional shaking. The exposure times were 0, 30, 60, 90 and 120 minutes. The UV irradiated cell suspension was stored in dark overnight, to avoid photo reactivation. Next, it was serially diluted in phosphate buffer and plated on nutrient agar plates with 1% shrimp shell powder. The plates were incubated for 24 hours at 37˚C and the numbers of colonies in each plate were counted. Each colony was assumed to have formed from a single cell. The strains were selected from the plates (0, 30, 60, 90 and 120 minutes UV exposure time) and tested for NK production. The survival rate was measured after every exposure time and the selected mutants were used for further studies (14).
3.7. Production of Nattokinase from the Mutant Strain
The Ultra violet-mutated strain was used for the production of nattokinase. One percent of seed inoculum was inoculated in 100 mL of optimized production medium in a 250 mL Erlenmeyer flask. The flasks were incubated for 24 hours at 37˚C. The culture was centrifuged at 8,000 rpm at 4˚C for 10 minutes and the clear supernatant was assayed for NK activity (15).
3.8. Partial Purification of Nattokinase
The ammonium sulphate precipitation was done at different saturations ranging from 20% to 80%. The precipitate was eluted in 20 mM Tris-HCl (pH 7).
3.9. Determination of Protein Content
The protein content of crude and partially purified enzyme extracts were determined by Lowry’s method (16).
3.10. Casein Digestion Assay
Nattokinase activity was determined using 0.1 mL of 2% casein, 0.7 mL of 0.1 M sodium phosphate buffer and 0.1 mL of partially purified enzyme of crude extract, incubated for 5 minutes at room temperature. The reaction was terminated by adding 0.1 mL of 1.5 M trichloroacetic acid (TCA) and centrifuged at 8,000 rpm for 10 minutes. Absorbance of the supernatant was measured at 560 nm. One unit of caseinolytic activity (U) was defined as the amount of enzyme releasing 1 µM of tyrosine equivalent/minute (8).
3.11. Blood Clot Lysis Assay
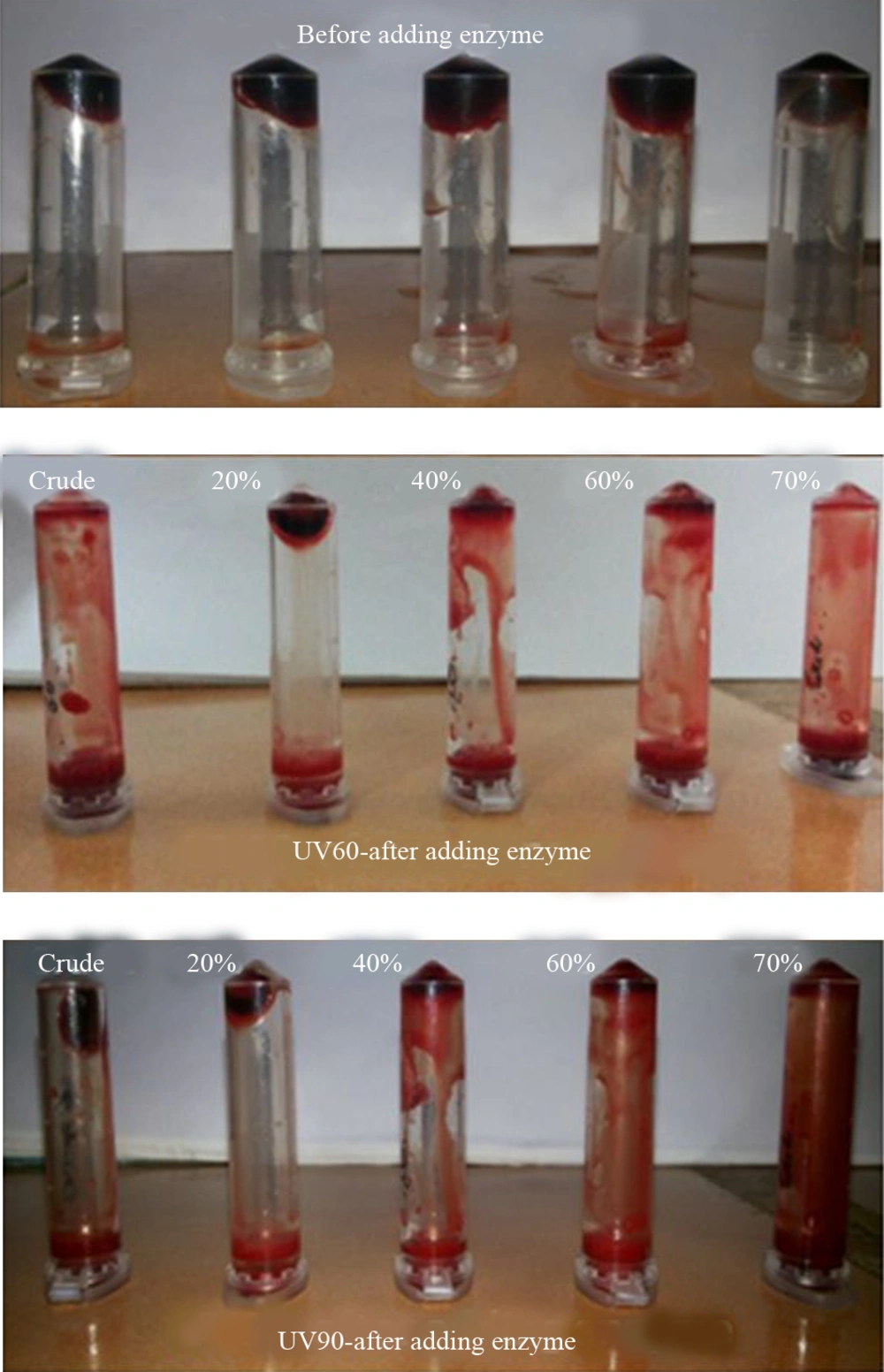
Venous blood drawn from healthy volunteers were transferred to different pre weighed sterile micro-centrifuge tubes (500 μL/tube) and incubated at 37˚C for 45 minutes. After clot formation, serum was completely removed (aspirated out without disturbing the clot formed) and each tube with clot was again weighed to determine the clot weight (clot weight was equal to weight of clot containing tube minus weight of tube alone). Each microcentrifuge tube, containing clot, was properly labeled and 100 μL of crude enzyme and ammonium sulfate precipitated enzyme was added to the clots. All the tubes were then incubated at 37˚C for 90 minutes and observed for clot lysis. After incubation, the obtained fluid was removed and the tubes were again weighed to observe the difference in weight after clot disruption. The obtained difference in weight taken before and after clot lysis was expressed as percentage of clot lysis (17, 18).
3.12. Fibrin Degradation Assay
The modified fibrin plate method was used for measuring the fibrinolytic activity of the crude enzyme and the partially purified enzyme of mutated strain (7, 19). Petri dishes containing 9 mL of 0.2% fibrinogen solution were placed on a horizontal glass plate. To each of these petri dishes, 0.2 mL of plasminogen (10 U) (Sigma Aldrich) was added and mixed well. Clotting was induced by the addition of 0.2 mL of thrombin solution (20 U). In order to speed up the clotting process, the plates were incubated at 37˚C for 15 minutes. Fresh Plates were prepared every time. A known quantity of the enzyme solution was placed as small droplets on the surface of the fibrin clot. The plates were then incubated at 37˚C for two hours and visually inspected for liquefaction.
3.13. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
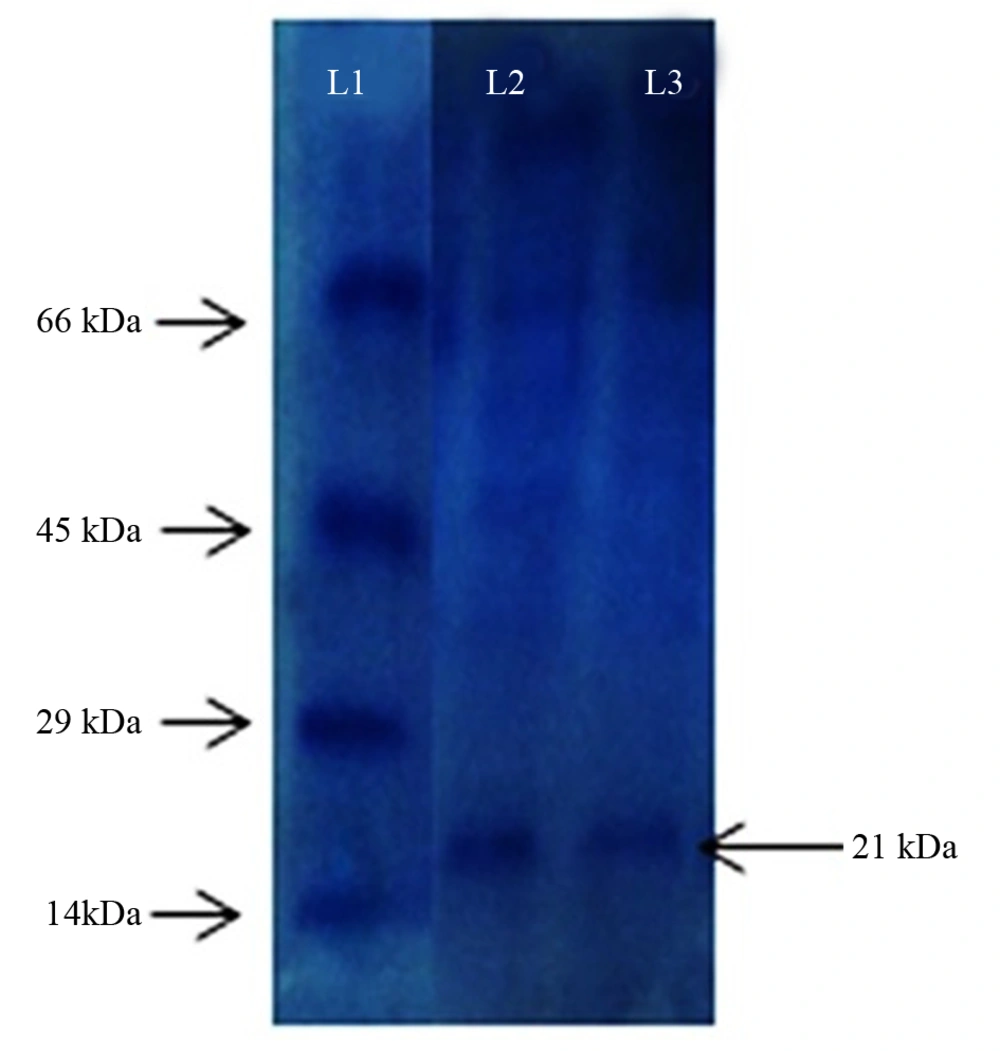
sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) was carried out to determine molecular mass and the purity of the partially purified enzyme, using 4% acrylamide in the stacking gel and 12.5% acrylamide in the separating gel. Protein bands were visualized by staining with Coomassie R 250 brilliant blue. Previous studies used phosphorylase b (molecular mass, 97.2 kDa), albumin (66.4 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa) and lysozyme (14.3 kDa) as the protein standards (20).
4. Results
4.1. Isolation and Screening
A total of 15 different cow milk samples were screened in this study. The isolates were selected based on β-hem olytic activity. Among 20 isolates, four isolates showed β-hemolytic activity. All these strains were further screened for NK production. Only one strain (CMSS) showed efficient hydrolysis in the casein hydrolytic assay.
4.2. Biochemical and Molecular Characterization
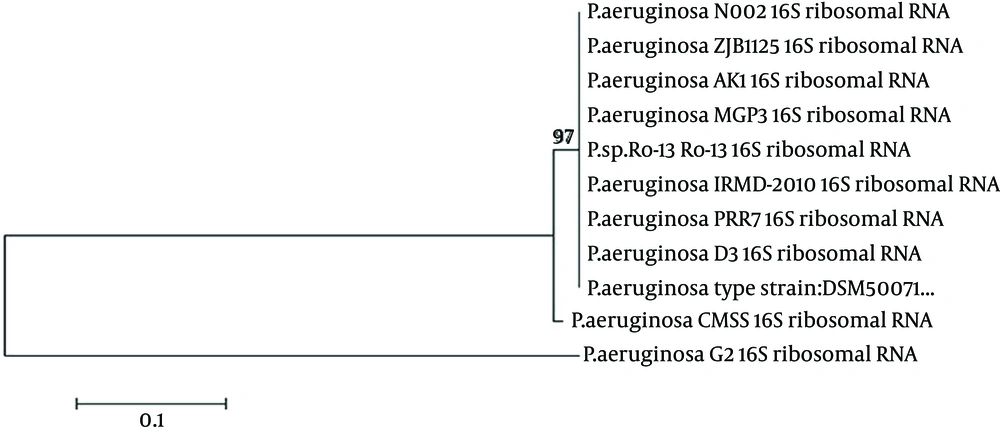
The CMSS isolate was found to be a motile and gram-negative rod. The biochemical characteristics of the isolate were as follows; catalase (+), Oxidase (+), Indole (-), Methyl red (-), Voges-Prausker (VP) (-), citrate utilization (+), urease (+), nitrate reduction (+), glucose (-), sucrose (-), lactose (-) and maltose (+). Based on the morphological and biochemical characterization, the CMSS strain was identified as Pseudomonas sp. The 16S rRNA sequencing was exported to the database and checked for homologous alignment. Based on the alignment results, the CMSS strain was confirmed as P. aeruginosa, which showed 99% similarity with other Pseudomonas sp. The partial 16S rRNA sequences were deposited in Gen Bank under the accession number JX112657. Aphylogenetic tree was constructed for the CMSS strain with bootstrap values (Figure 1). The phylogenetic tree based on 16S rRNA sequences showed that the isolate occupies a distinct phylogenetic position within the P. aeruginosa family. Based on the molecular taxonomy and phylogeny, the strain was identified as P. aeruginosa and designated as P. aeruginosa CMSS.
4.3. Effect of Medium Composition and Culture Conditions on Enzyme Production
In our preliminary experiments, the potent isolate Pseudomonas sp. CMSS in the production medium showed maximum enzyme productivity with maximum protein content (2.7 mg/mL) and enzyme activity (1045 U/mL). Different parameters, pH and temperature affect various biological processes and play important roles in the optimization process. The optimum enzyme activity was observed at pH 7 (1514 U mL-1 and 2.8 mg mL-1) and 25˚C (1532 U mL-1 and 2.8 mg mL-1). However, after a certain optimum percentage, the addition of further inoculum negatively affects fermentation by decreasing the dilution rate (Figure 2). Based on the obtained results, the optimum carbon source for the enzyme productivity of the P. aeruginosa CMSS strain was found to be 1% sucrose, in the presence of which maximum NK activity of 1721 U mL-1 was observed. Much less enzyme activity was obtained in the medium supplemented with glycerol (518 U mL-1). Furthermore, P. aeruginosa CMSS grown on shrimp shell powder supplemented medium, showed better activity (2524 U mL-1) than the strain grown in medium with other nitrogen supplements. The inoculum concentration of 1% was more favorable than the generally employed concentrations of up to 3%. The protein content and enzyme activity was found extremely high with the inoculum amount of 1% (2581 U mL-1). The production rate gradually increased with the increase in inoculum percentage (Figure 3).
4.4. Effect of Ultra-Violet Exposure on Survival Rate
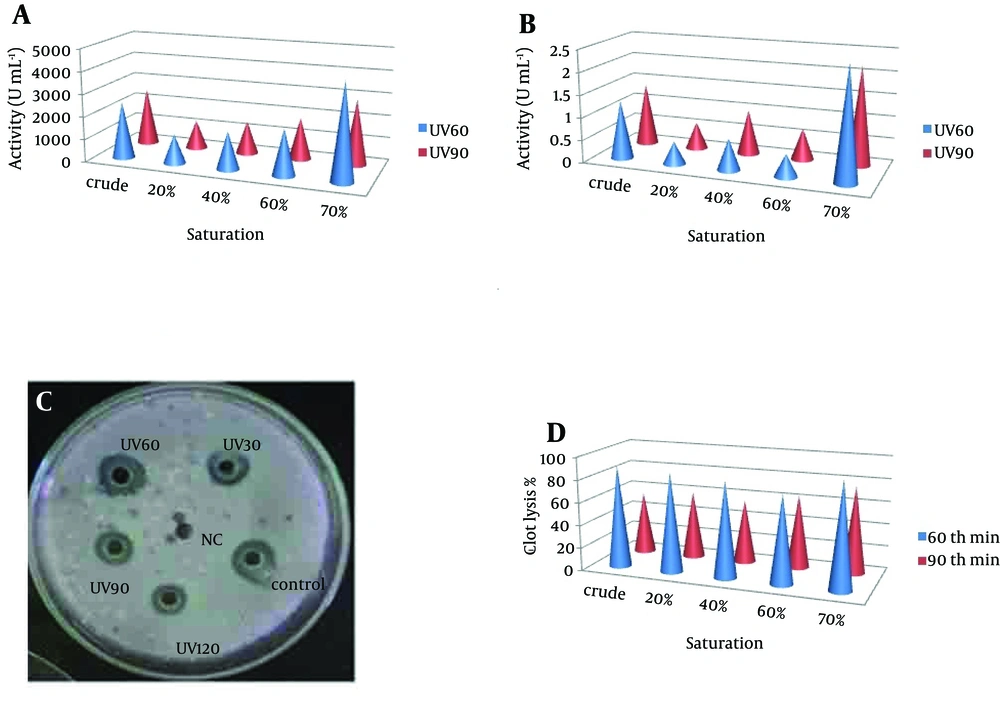
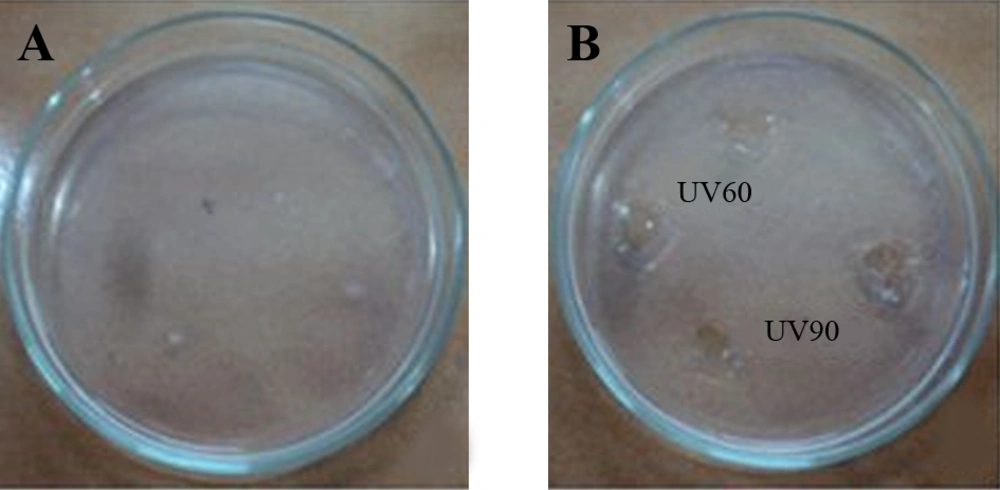
Pseudomonas aeruginosa CMSS was exposed to UV light for 30, 60, 90 and 120 minutes. Survival rate of parent strain was found to be inversely proportional to the exposure time (Table 1). Minimal survival rate of 44.1% was observed after exposure to120 minutes of UV-irradiation at 254 nm. The mutant colonies from UV60 and UV90 showed maximum zone of hydrolysis for NK by radial caseinolytic plate assay (Figure 4).
| No. of Colonies/Plate | Percentage of Survival Rate | Percentage of Death | |
|---|---|---|---|
| Control | 340 | 100 | 0 |
| UV30 | 280 | 82.35 | 17.6 |
| UV60 | 232 | 68.2 | 31.7 |
| UV90 | 210 | 61.7 | 38.2 |
| UV120 | 150 | 44.1 | 55.9 |
(a) Nattokinase Activity of Partially Purified Enzyme From UV60 and UV90 (b) Protein Content of Partially Purified Enzyme From UV60 and UV90 (c) Radial Caseinolytic Assay of Mutant Strains UV30, UV60, UV90 and UV120, and Control-Wild Strain and Negative Control (NC) (d) Percentage of Blood Clot Lysed After Addition of NK From Mutant UV60 and UV90
4.5. Nattokinase Activity
The NK activities of the UV60 and UV90 crude enzyme and ammonium sulphate precipitates of 20%, 40%, 60% and 70% were determined. The activity and protein of the 70% precipitate mutant UV60 strain expressed two-fold increase (4263U mL-1) when compared to the UV90 mutant (2568 U mL-1). The comparison of crude enzyme of mutant UV60 and UV90 expressed less activity than the partial purified. Similarly the protein content of mutant UV60 at 70% concentration was higher (2.5 mg mL-1) when compared to the mutant UV90 strain (2.3 mg mL-1) as well as the crude enzyme of wild strain (1.3 mg mL-1). Blood clot lysis activity was observed for the crude enzyme and ammonium sulphate precipitates of 20%, 40%, 60% and 70% NK enzyme from mutant strain UV60 and UV90. Furthermore, 94% clot lysis was visually observed after 10 minutes in the tube that received 70% precipitate of mutant UV60 strain while 77% clot lysis was observed in the tube that received70% precipitate of mutant UV90 strain (Figure 5). Fibrinolytic activity was determined by the in vitro fibrin plate method. The partially purified 70% precipitate NK enzyme from mutant UV60 strain showed maximum hydrolysis fibrin clot liquefaction when compared to NK enzyme from mutant UV90 strain, which was visually observed after two hours of incubation at 37˚C (Figure 6).
4.6. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
The molecular weight of the partially purified NK from UV60 was found to be 21 KDa, which was comparable by the banding patterns along with the protein marker. This confirms the NK produced by Pseudomonas sp. CMSS (Figure 7).
5. Discussion
The selection of suitable substrate is a significant factor for the enhanced production of enzyme. In the present research, the optimization studies for P. aeruginosa CMSS was done by varying physical and chemical parameters for maximum NK production. Nattokinase (fibrinolytic enzymes) was discovered from various microbes, which includes bacteria, actinomycetes, fungi, and algae (21). The optimization of culture conditions ensures ideal conditions for the growth and better production of enzymes in vitro. The production of NK has been enhanced by physical and chemical mutagenesis. The results of the present study are supported by existing reports on enhanced production of NK by UV-mutated Bacillus spp., which significantly demonstrated better activity compared to the wild type strain (22).
In the Holmstrom method, the extent of lyses was checked after the coagulation of blood, in order to determine enzyme activity. One unit of enzyme activity was defined as the minimum amount of enzyme at which blood clot lysis occurred completely (18, 22, 23). Thereby, it can be concluded from the results that 60 and 90 minutes of exposure time to UV was the most effective in producing favorable mutation that enhanced NK production with a higher activity. Many reports presented microbial nattokinase that had similar molecular weight as P. aeruginosa CMSS NK, included the fibrinolytic alkaline protease of Fusarium sp. BLB (27 kDa), the metalloprotease of C. militaris, the serine protease of Bacillus amyloliquefaciens DC-4 (28 kDa) and the metalloprotease of Armillaria mellea (21 kDa) (24-26).
Nattokinase produced from P. aeruginosa CMSS (Gen bank accession no. JX112657) was found to have more tendency to lyse blood clot. The efficacy of NK produced from wild and mutant strain showed varied difference in blood clot lysis. The optimized medium and culture conditions were used to cultivate UV-mutated strains, which significantly enhanced the production of nattokinase. Hence the results indicate that nutritional composition and culture conditions have important affects on fibrinolytic activity of NK from P. aeruginosa CMSS. Further studies should be focused on the biochemistry methods of improving the fibrinolytic activity of nattokinase, the molecular structure such as amino acid sequence, and secondary and tertiary structures.