1. Background
Hepatitis B virus (HBV), a member of the Hepadnaviridae family with partial double-stranded DNA, causes hepatitis B infection, which is as a major public health problem worldwide. Approximately 400 million people have been infected with the hepatitis B virus globally and are recognized as healthy carriers (1). Transmission methods of HBV include blood transfusion, saliva, semen, blood products, and tattooing (2, 3). Hepatitis B surface antigen (HBsAg) is an important diagnostic marker of HBV infection, but remains undetected in some patient sera (4). HBV replicates by reverse transcription of an intermediate RNA lacking proofreading activity, which results in the emergence of mutations in the HBV genome. Studies show that point mutations in the HBV genome reduce the secretion or synthesis of HBsAg or produce defective mutants. Moreover, deletion or nucleotide substitution is responsible for the occurrence of occult hepatitis B infection (OBI) (5).
Occult hepatitis B infection is characterized by the presence of HBV-DNA in the serum, without detectable HBsAg (6). The level of DNA in sera usually is < 104 copies/mL and can be detected by a sensitive PCR assay (7). Several reasons have been proposed for the presence of HBV-DNA in HBsAg negative patients, including integration of HBV-DNA into the host chromosome (8); genetic variations in HBV that suppress the expression of S gene (9); interaction of HBsAg with immune complexes, where HBsAg may be hidden (10); existence of a window after acute HBV infection; co-infection with hepatitis C virus; immunosuppression; or other host factors (11, 12). Conditions that weaken the immune system, such as AIDS or chemotherapy, can reactivate HBV in OBI patients and subsequently result in active or fulminant hepatitis B infection. Risk factors associated with hemodialysis patients include shared dialysis equipment, invasive procedures, and low response to HBV vaccination. Moreover, OBI causes the progression of conditions such as liver fibrosis, hepatocellular carcinoma (HCC), and fulminant hepatitis (13, 14). Investigation of these mutations will be helpful in predicting the progression of liver disease in hemodialysis patients with OBI.
2. Objectives
This study aims analyze mutations in the S and pre-core/core regions of HBV in dialysis patients with OBI in Ahvaz city, Khuzestan province, Iran.
3. Patients and Methods
In total, 216 hemodialysis patients, including 85 women and 131 men, were selected from the dialysis units of 4 hospitals in Ahvaz city from January 2012 to April 2012. Blood samples were collected and their sera were tested for HBsAg by ELISA (RPC/ Russia), according to the manufacturer’s instruction. In total, 13 sera samples collected from eight males and five females tested positive for HBsAg (RPC Diagnostic systems, Nizhny Novgorod, Russia) and were excluded from this study. The remaining 203 sera samples were tested for the presence of HBV-DNA and HBcIgG ELISA kit (DIA.PRO, Milan, Italy).
3.1. Genome Extraction
Genome extraction was conducted for 203 HBsAg-negative samples by High Pure Viral Nucleic Acid Kit (Roche/Germany) according to the manufacturer’s instruction.
3.1.1. The Nested-PCR for S Region
The following primers were used for detection and mutation analysis of the S region (High Pure Viral Nucleic Acid Kit (Roche, Indianapolis, USA): outer primers, SA1: 5′-ATCGCTGGATGTGTCTGCGG-3′ ′, and SA2: 5′-GGCAACGGGGTAAAGGTTCA-3′ (position 369–388 and 1136–1155); and inner primers, SB1: 5′-TTAGGGTTTAAATGTATACCC-3′ (position 822 - 842), and SB2: 5′-CATCTTCTTGTTGGTTCTTCTG-3′ (position 427 - 448) (15). First round PCR was performed with a 25 µL total volume, containing 7 µL of HBV-DNA as template, 2.5 µL 10x reaction buffer, 0.75 µL MgCl2 (50 mM), 0.5 µL forward/reverse primer, 1 µL dNTP (10 mM), 0.2 Taq DNA Polymerase (Cinna Gen, Tehran, Iran) (5 u/µL), and 13.55 µL of double-distilled water. The thermal cycler was programmed for the first round as follows: 35 cycles with initial denaturation at 94°C for 5 minutes, 94° C for 1 minute, 54°C for 1 minute, 72°C for 1.5 minutes, and final extension at 72°C for 10 minutes. The second round was conducted with the same components except for the DNA template; 5 µL of first round product was used as the template for second round. The second round was programed as follows: 30 cycles with initial denaturation at 94°C for 5 min, 94° C for 1 minute, 45°C for 1 minute, 72°C for 1 minute, and final extension at 72°C for 10 minutes. The 416 bp PCR product was subjected to electrophoresis on a 2% agarose gel supplemented with DNA safe Stain agarose and DNA Safe stain (Cinna Gen, Tehran, Iran) and visualized using a UV-trans illuminator.
3.1.2. Nested PCR for Pre-Core/Core Region
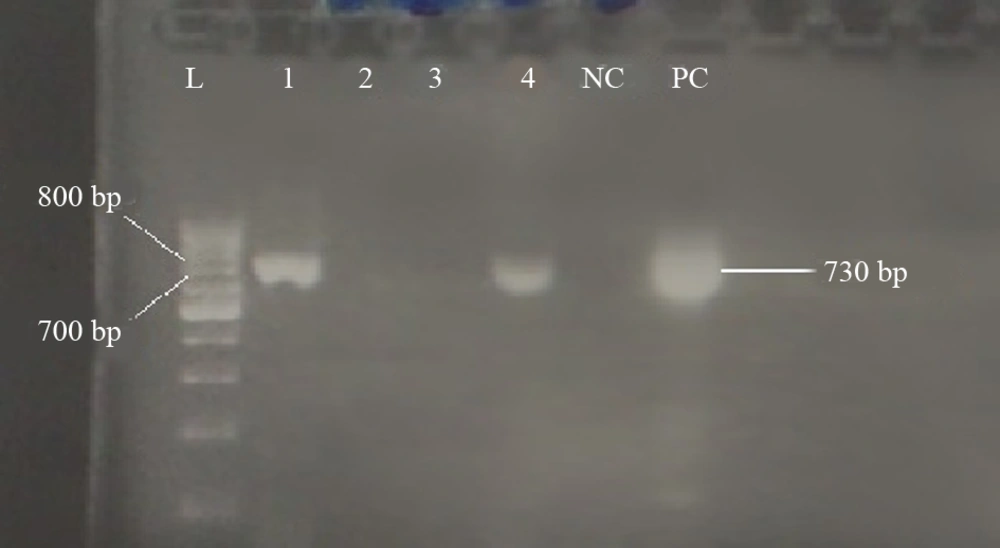
The following primers were used for detection of pre-core/core mutations UV-trans illuminator (Kiagen, Tehran, Iran): outer primers, PC16: 5′-GTTGCATGGAGACCACCGTGAAC-3′ (nt. 1605–1627) and PC17: 5′-CTTCTGCGACGCGGCGATGGAGA-3′ (nt. 2410–2432); and inner primers, PC70: 5-′CATAAGAGGACTCTTGGACT-3′ (nt. 1655 - 1674) and PC74: 5′-GGCGAGGGAGTTCTTCTTC-3′ (nt. 2378 - 2396) (15). First round PCR was performed with 25 µL volume, containing 7 µL of HBV-DNA as template, 2.5 µL 10x reaction buffer, 0.75 µL MgCl2 (50 mM), 0.5 µL forward/reverse primer, 1 µL dNTP (10 mM),0.2 Cinna Gen Taq DNA Polymerase (5 u/µL), and 13.55 µL of double-distilled water. The reaction mixture was subjected to thermal cycler (TC-512, Techne, Staffordshire, UK) with the following program: 35 cycles with initial denaturation at 94°C for 5 minutes, 94°C for 1 minute, 60°C for 1 minute, 72°C for 2 minutes, and final extension at 72°C for 10 minutes. The first round of PCR product was used as the template for the second round. The amount of PCR components was the same as the first round except for the DNA template, which was 5 µL in the second round. The thermal cycler was programmed as follows: initial denaturation at 94°C for 5 minutes, 30 cycles consisting of 94°C for 1 minute, 44°C for 1 minute, 72°C for 1.5 minutes, and final extension at 72°C for 10 minutes. The 730 bp PCR product was subjected to electrophoresis on a 1% agarose gel supplemented with DNA safe Stain (CinnaGen) and visualized using a UV-trans illuminator.
3.2. TA Cloning
The TA cloning was performed for positive PCR products of S and pre-core/core regions. The purified PCR products of both S and pre-core/core regions were cloned separately with InsTAclone PCR Cloning Kit (Thermo Scientific/USA) according to the manufacturer’s instruction. The recombinant vectors were transformed into the Escherichia coli TOP 10 strain. After blue-white screening, two or three colonies were selected and tested by colony PCR. Plasmid extraction was performed for the detection of positive PCR colonies by using AccuPrep Plasmid MiniPrep DNA Extraction Kit (Bioneer/Korea) based on the protocol and sequenced using a universal primer (M13-20).
3.3. Mutation Analysis of HBV S and Pre-Core/Core S Regions
Sequencing of both nested PCR products and TA cloning products of S and pre-core/core regions was performed by Bioneer (Korea). The results were analyzed by MEGA 5.2, Gene Runner, and web-based tools such as NCBI, Hepatitis B Virus Database, and Geno2pheno. The reference sequence from GeneBank with accession number NC-003977.1 was used to compare and detect the presence of mutations in isolated HBV samples from hemodialysis patients.
4. Results
This study involved 203 hemodialysis patients who tested negative for HBsAg, including 123 (60.6%) males and 80 (39.4) females with an average age of 51 ± 15.37 years. PCR results revealed that out of 203 samples, 175 (81.01%) were negative for hepatitis B core antibody (HBcAb), six (2.9%) were positive for HBV-DNA including four (1.97%) for S (Figure 1) and two (0.98%) for pre-core/core regions (Figure 2) and were recognized as OBI (Table 1). Out of six positive HBV-DNA samples, four (1.97%) samples were positive for the S region. All positive PCR samples were negative for both HBeAb and HBeAg. PCR analysis of the HBV-DNA of the S region showed that all four samples had “YMDD” motif in position 204 (rtM204), which revealed Lamivudine sensitivity. Moreover, sequencing of HBV-DNA of the S region of all four (1.97%) samples showed arginine (R) in position 122 and lysine (K) in position 160, indicating the presence of HBV subtype awy and genotype D.
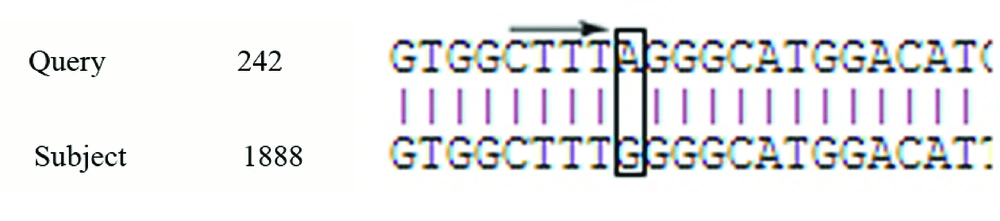
In this study, we observed a substitution in the RT region (Y135S) and three amino-acid substitutions in the S region (T127P, P153L, F170S); T127P was an escape mutant that produce different surface antigen (Figure 3). There was 100% homology among all four samples in the S region. One (0.4%) of the six OBI patients was HBcAb positive, which is known as seropositive occult hepatitis B infection. Analysis of the pre-core/core regions showed nucleic acid replacement in position G1896A, which results in an unwanted stop codon ( Figure 4).
| Variable | HbsAg/PCR + | HbsAg/PCR - |
|---|---|---|
| Gender | 2 F and 4 M | 51 F and 82 M |
| Age, y | 59 ± 19.5 (38 - 77) | 51 ± 15.37 (10 - 82) |
| Receiving blood product | 3/6 | 95/133 |
| Tattooing | 2/6 | 30/133 |
| Surgery | 4/6 | 103/133 |
| HBV pre-core/core region positive | 2/6 | - |
| HBV S region positive | 5/6 | - |
| ALT, IU/L | 4.08±6.45 | 9.8 ± 10 |
| AST, IU/L | 9.5±6.95 | 16.3 ± 12.5 |
| Total Bilirubin, mg/dL | 0.43±0.1 | 0.63 ± 0.18 |
| Direct Bilirubin, mg/dL | 0.16±0.08 | 0.33 ± 0.13 |
| Alkaline Phosphatase, IU/L | 81±236.03 | 364.37 ± 212.2 |
a Abbreviations: F, Female; M, Male.
5. Discussion
Occult hepatitis B infection is a major health concern globally, and a major cause of chronic liver diseases, cirrhosis, and infections transmitted by blood transfusion (16). Moreover, OBI diagnosis is possible by analyzing low levels of the harbored HBV-DNA (less than 200 IU/mL) by PCR, when HBsAg is absent (17, 18). Several studies have shown that OBI increases the risk of HBV transmission through blood transfusion in hemodialysis patients (19, 20); however, the prevention of OBI transmission in hemodialysis patients remains a challenge (21). There is notable variance in the prevalence of OBI in hemodialysis patients, ranging from 3.11% to 58%, worldwide (3, 22, 23). This study reported the presence of OBI in 2.9% of hemodialysis patients. Studies show that antibody against a determinant (amino acid 120 - 150) of the surface antigen is more important for immunity against HBV infection, and mutations in this region can create escape mutants and cause OBI (24-26). For example, Jeantet et al. found mutations in the S region of HBV genome (27). Minuk et al. reported that out of 239 HBsAg negative patients, nine (3.8%) were HBV-DNA positive and seven of the nine (78%) showed mutations in the S region (G145R) (28). In another study, Mu et al., reported that out of 46 HBsAg negative children, five children had OBI and a sequence analysis of the S region showed mutation in position C139S, which makes HBV vaccine inactive (29).
Substitution of amino acid 145 is a frequent mutation in the S region, which leads to the production of different surface antigen and escape mutants. Moreover, positions such as 144, 143, 142, 141, 137, 133, 120, 124, 126, and 129 can be affected by mutations (24). Awerkiew et al. determined three different S gene mutations (L109R, C137W, G145R) in a patient with occult hepatitis B, which led to false negative results for HBsAg but positive for HBsAb (30). Nainan et al. reported that the T127R mutation is an escape mutant in the S region (24). We observed a substitution in the 127 position, but in our study, T was replaced with P instead of R, which is not a frequent substitution in this position. This result is similar to that reported by Sayan et al, who also reported a T to P substitution at position 127 (26). Hou et al. reported 32 amino acid substitutions between positions 100 and 160 of the MHR (major hydrophobic region) of HBsAg, which were observed in OBI patients (20, 31).
In our study, we detected a new substitution in position 153 (P153L), which can create escape mutant when placed in MHR (20). Van Hemert et al. have identified a deletion mutation in RNA splicing stage that delete nucleotides between positions 2986 and 202, which blocks 274 surface protein gene expression without affecting polymerase, core, and X-protein functions (32). Mutations in the pre-core region can create a stop codon that prevents production of the pre-core protein and HBeAg, which generally occurs in HBV genotype D (33). Moreover, Bremer et al. found an occult HBV with a stop mutation in the pre-core region of blood donors, which led to the loss of HBeAg expression (34). In our study, an unexpected stop codon was found in position 1896 of the pre-core region, which prevented the production of HBeAg in six OBI patients. All these patients tested negative for HBeAg and HBeAb. All four positive samples were genotype D, which was in accordance with other studies reported in Iran (1).
PCR revealed that two (0.98%) of the occult hepatitis B patients tested negative for the S region, while they tested positive for the HBV pre-core/core region. In a similar study, Wilson et al. reported the absence of positive PCR results for the pre-core region in six patients, who exhibited positive PCR results for S region (35). The negative PCR result in the pre-core region is because of a mutation in the 1896 position, which has a significant relationship with viral load (36). This study demonstrated a new substitution in position 153 of the HBV genome, which creates an escape mutant HBV in OBI patients. Hence, whole genome sequencing is important and useful for recognizing other mutations in other regions of the HBV genome to detect new escape mutants in OBI patients in southwestern Iran.