1. Background
Blastocystis hominis is a common intestinal protozoan parasite of humans and many animals (1, 2). This parasite is perhaps the most common eukaryotic organism in the human digestive system (3), with a prevalence of up to 60% reported in some developing countries (4). The parasite’s prevalence in developed countries, on the other hand, is reportedly 5% - 20% (1). Standards of health care, garbage disposal, as well as food and water contamination must be improved in developing countries to lower the parasite prevalence (5). Blastocystis spp. are zoonotic parasites and do not have a specific host (2, 6). Blastocystis transmission occurs primarily by fecal-oral means, which is exacerbated in poor sanitary conditions (7). Contaminated water is the most important issue in spread of the parasite (8). Despite extensive research, there is no consensus regarding the pathogenesis of Blastocystis spp. Some of the symptoms that have been attributed to Blastocystis are nausea, anorexia, abdominal pain, bloating, and acute or chronic diarrhea (9-12).
Blastocystis spp. have great genetic diversity (13, 14). Based on molecular analysis, Blastocystis isolates from humans, mammals, and birds can be assigned to nine subtypes (15). Differentiation of Blastocystis species by routine methods is not possible, and DNA-based methods are therefore essential for the detection of genetic variation in these organisms. Molecular epidemiological studies are particularly useful in ascertaining transmission patterns, host specificity, and chemotherapeutic drug resistance. The prevalence and genotype diversity of Blastocystis spp. in Iran have not been studied extensively. Previously, PCR-RFLP and PCR with seven pairs of sequence-tagged site (STS) primers were used to assess the genetic diversity of Blastocystis spp. in Iran (16, 17), and in this study, specific PCR products were sequenced for further assessment of genetic diversity among Blastocystis spp. in Iran.
2. Objectives
The aim of this study was to investigate the prevalence and genotype distribution of Blastocystis spp. among the Baghmalek people in southwestern Iran.
3. Materials and Methods
3.1. Study Area
Baghmalek city is located 120 km from the Ahvaz metropolitan in southwestern Iran. The city has a population of 110,000 of which more than 80% live in rural areas.
3.2. Sample Collection and Microscopic Diagnostics
Over a period of 3 months, after study subjects completed questionnaires, 1,410 stool samples were collected from patients from five districts of Baghmalek city: north, south, east, west, and central. Patients completed questionnaires and provided written consent. Stool samples were used for two fecal smear tests: normal saline and Lugol’s iodine smears. Formalin ethyl acetate concentration and trichrome staining methods were also performed (1). Approximately 200 mg of each positive sample was stored at -20˚C until subsequent molecular analysis.
3.3. PCR Amplification and Sequencing
DNA was extracted from fecal specimens by using an AccuPrep® Stool DNA extraction kit (Bioneer, Daejeon, South Korea) according to the manufacturer’s instructions. The specific primers used to amplify the SSU rDNA gene from Blastocystis-positive specimens were Blast 505 - 532 (5′ GGA GGT AGT GAC AAT AAATC 3′; forward) and Blast 998 - 1017 (5′ TGC TTT CGC ACT TGT TCATC 3′; reverse) (18). PCR amplifications were performed using AccuPower® PCR PreMix, a ready-to-use PCR reagent (Bioneer), where each reaction contained 7 µL distilled water, 2 µL of each primer, and 4 µL extracted DNA. The PCR conditions were as follows: an initial pre-heating step at 95˚C for 4 minutes; cycles of 95˚C for 30 seconds, 54˚C for 30 seconds, and 72˚C for 30 seconds; and a final extension step at 72˚C for 5 minutes. Amplicons were visualized with agarose gel electrophoresis (1.5% agarose), and samples yielding products of 500 (479) bp were considered positive for Blastocystis. Positive and negative controls were included and PCR products were sent to South Korea for sequencing.
3.4. Sequence Analysis
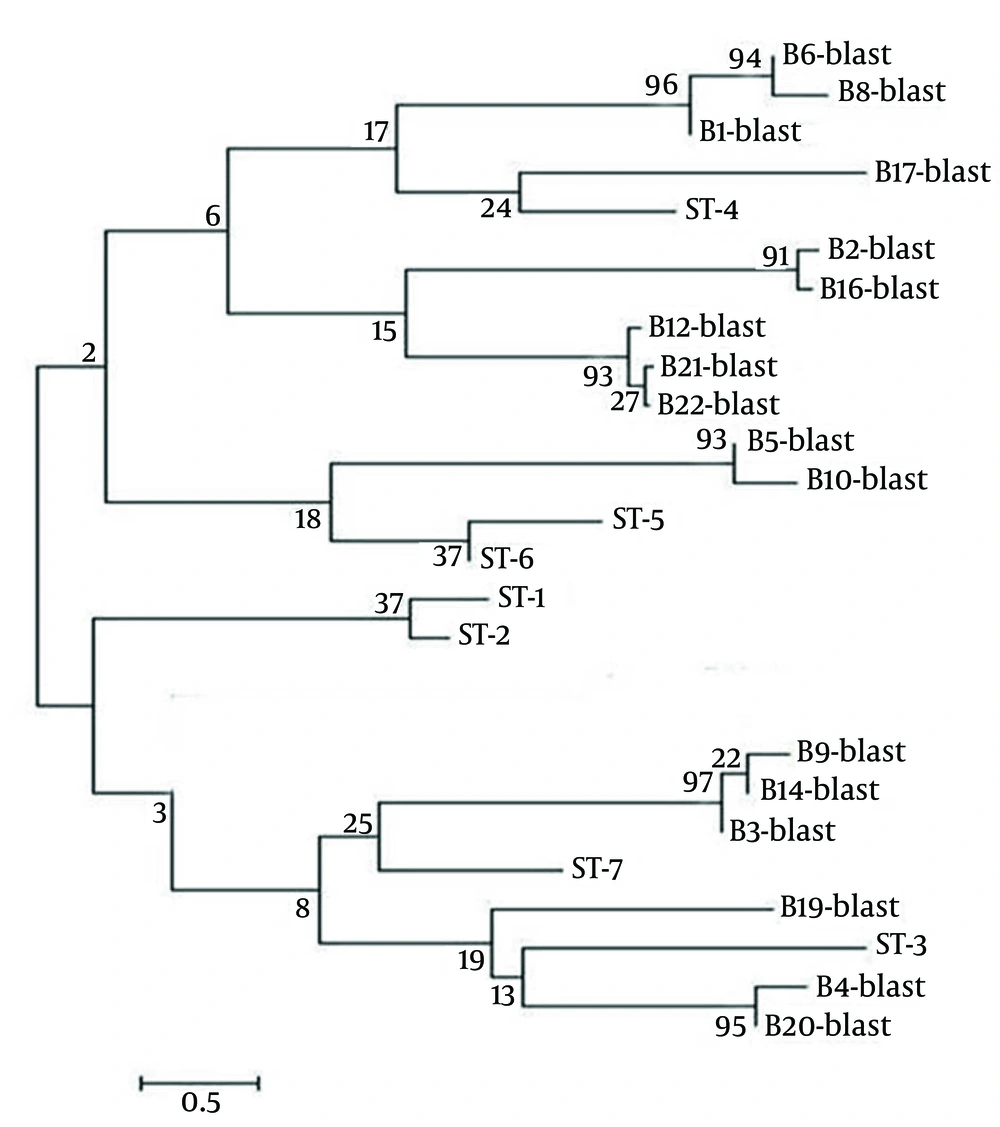
DNA sequence analysis was performed on 22 PCR-positive samples. The SSU rDNA sequences obtained were compared with those available in GenBank by using the BLASTN program (http://www.ncbi.nlm.nih.gov/BLAST/). ST-1|HQ641595, ST-2|HQ641605, ST-3|HQ641611, ST-4|HQ641621, ST-5|HQ641630, ST-6|HQ641658, ST-7|HQ641661. Distance-based analysis was conducted on the sequencing data by using molecular evolutionary genetic analysis version 4 (MEGA 4) (19) and a phylogenetic tree was constructed using the neighbor-joining method (20) with the Kimura 2-parameter model (21). The support of monophyletic groups was assessed by the bootstrap method with 1,000 replicates (22).
3.5. Statistical Analysis
Frequency distributions of strains and demographic information were analyzed by descriptive statistical methods, and qualitative variables were analyzed by the chi-square test.
4. Results
The prevalence of Blastocystis spp. based on microscopic analysis was found to be 3.33% among people referred to Baghmalek medical centers. Of 1,410 patients whose stool samples were examined by microscopy, 602 (42.7%) were males, 22 of whom were infected with Blastocystis spp., and 808 (57.3%) were females, 25 of whom were infected (age range: 6 - 60 years). Although the infection rate was greater in males than in females, there was no significant statistical association between Blastocystis infection and gender (P = 0.57; Table 1). The highest positivity rate (4.51%) was observed in the group of subjects less than 15 years old, but statistical analysis indicated no significant association between Blastocystis infection and age (P = 0.3; Table 2). Of 47 samples characterized as Blastocystis-positive by direct microscopy methods, 25 did not yield Blastocystis-specific PCR products.
| Gender | No. Examined | Positive Cases |
|---|---|---|
| Male | 602 | 22 (3.65) |
| Female | 808 | 25 (3.09) |
| Total | 1,410 | 47 (3.33) |
a The values are presented as No. (%).
| Age, y | No. Examined | Positive Cases |
|---|---|---|
| 0 - 15 | 487 | 22 (4.51) |
| 16 - 30 | 544 | 10 (1.83) |
| > 30 | 379 | 15 (3.95) |
| Total | 1410 | 47 (3.33) |
a The values are presented as No. (%).
Seventeen samples found to be Blastocystis-positive by PCR were sequenced and Blastocystis subtype analysis yielded the following results: subtype 4 was the most predominant subtype, infecting nine (40.9%) individuals, while subtypes 3, 5, and 7 were isolated from only three (13.6%), two (9.1%), and three (13.6%) cases, respectively (Table 3). It should be noted that, despite specific PCR products being obtained, the subtype(s) of five sequenced samples could not be determined.
| Type | Number | Percent |
|---|---|---|
| ST3 | 3 | 13.6 |
| ST4 | 9 | 40.9 |
| ST5 | 2 | 9.1 |
| ST7 | 3 | 13.6 |
| Unknown | 5 | 22.8 |
| Total | 22 | 100 |
A phylogenetic tree of the Blastocystis isolates is displayed in Figure 2. The nucleotide sequences of the 21 sequenced and subtyped samples from this study have been deposited in GenBank under accession numbers AB915194 - AB915214.
The phylogenetic tree was inferred using the neighbor-joining method. Bootstrap proportions (%) are attached to the internal branches (1,000 replicates). Names of the specimens, compared subtypes (ST) and GenBank accession numbers are as follows: B4-blast, BMBlasto4|AB915198; B19-blast, BMBlasto19|AB915208; B3-blast, BMBlasto3|AB915205; B9-blast, BMBlasto9|AB915201; B14-blast, BMBlasto14|AB915199; B5-blast, BMBlasto5|AB915207; B10-blast, BMBlasto10|AB915209; B6-blast, BMBlasto6|AB915197; B8-blast, BMBlasto8|AB915196; B1-blast, BMBlasto1|AB915202; B17-blast, BMBlasto17|AB915210; B12-blast, BMBlasto12|AB915194; B21-blast, BMBlasto21|AB915213; B22-blast, BMBlasto22|AB915214; B2-blast, BMBlasto2|AB915211; B16-blast, BMBlasto16|AB915203. The nucleotide sequences of B20-blast was not deposited in GenBank.
5. Discussion
In this study Blastocystis was detected in 3.33% of stool specimens examined by microscopy, which is in agreement with the prevalence (2.2% - 28.2%) of Blastocystis previously reported in microscopy-based studies in Iran (23, 24). The prevalence of this parasite determined by traditional parasitological techniques is lower than that determined by PCR- and cultivation-based studies (25, 26). This discrepancy can be attributed to the resemblance of Blastocystis with yeast, Cyclospora spp., and fat globules. The prevalence rate of infection in the current study was comparable to that reported in developed countries, which may be because of acceptable primary health and environmental conditions in the restricted area.
In this study, the highest prevalence rate of infection was observed in the ≤ 15 year age group. This may be because of lack of parental supervision or lack of personal hygiene in this age group, increasing the susceptibility of these subjects to infection. Higher parasite prevalence was observed among males in this study compared with females, which is consistent with the findings of similar studies in Libya (27) and Turkey (28), which also reported higher rates of infection in males than in females. The differences in the prevalence rates of Blastocystis infection with age (P = 0.3) and gender (P = 0.57) in this study were not statistically significant, and thus, it seems that age and sex are not risk factors for Blastocystis infection.
This is one of few studies to date investigating the molecular epidemiology of Blastocystis infections in Iran. Subtype 4 (ST4) of Blastocystis was identified in the current study, and in two studies on Blastocystis previously conducted in Iran, ST4 was not reported (15, 16). The subtyping in these previous studies was conducted by PCR-RFLP, which is not considered an accurate approach for Blastocystis subtype identification, because the same RFLP profile does not always represent the same sequence (29-31) and sequence-tagged site primers of some genotypes cannot be detected by this method (32). In the present study, specific PCR product sequencing was used instead, yielding results similar to those reported by Poirier et al. (33) in France and Stensvold et al. (34) in Denmark. Sequencing was used for subtyping in both these studies. Noel et al. (35) and Silberman et al. (36) believe that rodents are reservoirs host of ST4, and high prevalence of ST4 in the study area may therefore be associated with rodent infection. This potential relationship, however, requires further investigation for confirmation.
The results of this and other studies demonstrated significant variation in Blastocystis strain prevalence within populations: ST3 has been reported to be predominant in several studies (37-39), while it was not very prevalent (13.6%) among the positive isolates in the current study. ST5, which was previously reported at a low rate in humans (5), was found to have a prevalence of 13.6% among positive samples in this study. Although there is no clear explanation for these differences, they may be a result of differences in lifestyles as well as differences in the animal reservoirs in different study areas. In the current study, ST7 was identified, like in other studies in the Middle East (40, 41). Other known Blastocystis subtypes including ST1, ST2, ST6, ST8, and ST9 were not identified in our study. Larger sampling in further studies may help to explain this finding. As also observed in other studies (16, 38, 41), some samples (n = 25) found to be Blastocystis-positive by direct microscopy in this study did not yield Blastocystis-specific PCR products. This may be due to a number of factors including instability in sampling, improper storage and transportation, feces inhibitors, and low concentration of parasites (42-44).
This study was an assessment of the prevalence and subtypes of Blastocystis in Baghmalek in southwestern Iran. Unlike many studies conducted in the Middle East and Far East, ST4 was found in this study to be the most common subtype a discrepancy that requires further investigation. As previously reported in other studies, ST3, ST5, and ST7 were also identified in our study. Further investigation into the prevalence and subtypes of Blastocystis in this and other study areas should include investigation into routes of transmission and identification of zoonotic subtypes.