1. Background
Aflatoxins are secondary metabolites produced by species of Aspergillus, mainly A. flavus and A. parasiticus. Aflatoxigenic Aspergillus spp. can colonize a variety of substrates under a wide range of environmental conditions. Some aflatoxins can be highly carcinogenic or toxic to mammals and birds (1). The highly hepatotoxic aflatoxin B1 (AFB1) is of the greatest concern, since it can contaminate a wide range of agricultural products (2). Detection of AFB1 can result in great losses in agricultural income and has a significant negative economic impact. According to the FAO, 25% of the world food-crops are affected (3). A number of methods are developed for pre- and post-harvest aflatoxin management (4). Various clay-binders are proposed as food additives to inhibit AFB1 absorption from food stuffs (5). However, these approaches do not completely remove AFB1 safely, in a cost-effective manner. Thus, there is a need for a more effective method to detoxify AFB1-contaminated products.
AFB1 is impervious to degradation by high temperatures (6), which poses a problem for its industrial degradation. Other existing non-biological methods for AFB1 detoxification, by treatment with various physical or chemical agents (7), are not ideal in terms of safety, economy, and the quality of the treated products. Alternatively, biological AFB1 degradation, using plant extracts or various live organisms possessing AFB1-degrading activity and/or inhibiting the growth of A. flavus, is promising for decontamination. Numerous studies report the inhibitory properties of some fungi towards aflatoxigenic Aspergillus species and the microbial detoxification of AFB1-contaminated products by soil bacteria and fungi (8-12). At the same time, use of such microbial degradation can have disadvantages such as production of undesirable metabolites, poor flavor, or lowering nutritional value and acceptability of the products. However, isolation of microbial AFB1-catabolizing enzymes would avoid the drawback of decontamination by living microorganisms.
Despite the fact that many macromycetes and microorganisms were reported to degrade AFB1 under the laboratory conditions, no system is commercialized. Hence, researches are conducted to find an efficient method to degrade AFB1. In nature, A. flavus may produce AFB1 to out-compete other microorganisms. However, microorganisms that can share ecological niches with A. flavus could presumably survive as a result of secreting AFB1-catabolizing enzymes. Such niche-sharing microorganisms could serve as potential sources of aflatoxin biodestructors. This potential was demonstrated in our previous study (13), wherein various identified micromycetes, collected from natural substrates inhabited by A. flavus, possessed AFB1-degrading activity of the fungus itself or its culture liquid filtrate (CLF). The best results were observed for CLFs of P. glomerata and P. exigua (degradation of 66% and 99% of added AFB1 within 72 hours, respectively). However, P. exigua was excluded from the further studies due to its unstable AFB1-degrading activity.
2. Objectives
The current survey aimed to study the AFB1-degrading metabolites, produced by Phoma glomerata PG41, isolated from a natural substrate inhabited by aflatoxigenic A. flavus, and the preliminary determination of the nature of these metabolites.
3. Materials and Methods
3.1. Reagents
Solvents, chemicals, and AFB1 standard used in this study were purchased from Sigma Aldrich Corp., USA.
3.2. Strain and Culturing Conditions
Phoma glomerata PG41 was collected from the stored beans of Phaseolius vulgaris L., cultivated in and stored at the All-Russian Research Institute of Phytopathology (ARRIP, Moscow region). The strain was isolated by the serial dilution technique (14) and identified (15). The strain was maintained in the ARRIP Physiological Plant Pathology Laboratory on PDA at 4ºC. To obtain a submerged PG41 culture, a fermentation medium (13) was inoculated with spore suspensions of 14-day-old PG41 from PDA slants, grown in darkness at 25-27ºC, then incubated for seven days on Orbital Refrigerated Shaker E-25R, (New Brunswick, USA) at 27°С and 200 rpm.
3.3. Preparation of Culture Liquid Filtrate and Cell-Free Extract of Fungal Mycelium
The submerged PG41 culture was centrifuged (10'000 g, 4ºC, 30 minutes) to separate the mycelium and culture liquid. To obtain CLF, the supernatant was filtered through a Miracloth (Calbiochem®, Behring Diagnostics, USA), lyophilized, and dissolved in 100 mM Tris-HCL buffer (PH 8.3) containing 20 mМ EDTA-Na, 10 µM pyridoxal-phosphate, 1 mM phenylmethanesulfonyl fluoride, and 0.65 mМ dithiothreitol. To obtain cell-free extracts, mycelia were homogenized in the same Tris-HCl buffer (w/v 1:2) using an Omni-Mixer homogenizer (USA) at 10'000 rpm for 10 minutes and centrifuged (10'000 g, 4ºC, 30 minutes). The supernatant was saved and combined with a second supernatant collected from re-centrifugation of the cell debris from the first centrifugation.
3.4. Preparation of High-Molecular-Weight Protein-Enriched Fractions From the Cell-Free Extract and Culture Liquid Filtrate
High-molecular-weight (HMW) metabolites of P. glomerata were obtained from the cell-free extract and CLF by ultrafiltration using Ultracel® membranes (Molecular Weight Cutoff 3 kDa) combined with Amicon Ultra centrifugal devices (Millipore, USA). The fraction of ultrafilter-retained metabolites was adjusted to initial volumes to remove the accompanying concentration effects. Protein-enriched fractions of HMW metabolites were obtained by precipitation with ammonium sulfate (70% saturation, 4ºC, 18 hours), followed by centrifugation (10000g, 4ºC, 30 minutes), dissolving the pellet in extracting buffer (see 3.3), and overnight dialysis against this buffer. The obtained protein-enriched fractions were sterilized through 0.22-µm filters (Millipore, USA).
3.5. Aflatoxin B1 Degradation Assay
AFB1 was dissolved in 95% methanol and added to 1mL samples of sterile initial CLF and protein-enriched fractions of HMW metabolites to a final concentration of 2 µg/mL. Extracting buffer or nutrient medium, containing the same AFB1 concentration, served as corresponding controls. All samples were incubated for 72 hours at 27°С under aseptic conditions, as found appropriate for all discovered AFB1-destructing micromycetes (13). Each variant was tested in five replications.
3.6. Residual AFB1 Quantitation
Quantitation of residual AFB1 included extraction and HPLC techniques (16, 17), with modifications specified below. Residual AFB1 was extracted with chloroform on a shaker (200 rpm, 1 hour) followed by overnight incubation (no shaking, 4ºC). The chloroform phase was collected, and the water phase was twice re-extracted with chloroform. Chloroform extracts were combined and dehydrated with anhydrous Na2SO4. AFB1 was purified following elution from Al2O3 column by chloroform. Chloroform was removed by rotary evaporation (40ºC) and the residue was dissolved in 50% aquatic methanol (mobile phase). AFB1 was quantified by HPLC using a Waters 1525 Breeze HPLC system equipped with a Waters 2487 UV detector (Waters Corp, USA). Samples (5 µL) were applied to a temperature-controlled (27ºC) Diaspher 110-C18 column (6µm, 4 × 150 mm, Biochemack, Russia). AFB1 was eluted by the mobile phase (0.5 mL/min) and detected at 362 nm. The toxin concentration was calculated using a calibration curve based on an AFB1 standard. All samples were analyzed in triplicate. The data were analyzed using STATISTICA 6.0 software, and means of triplicates ± standard deviations were reported. P ≤ 0.05 was considered as level of significance.
4. Results
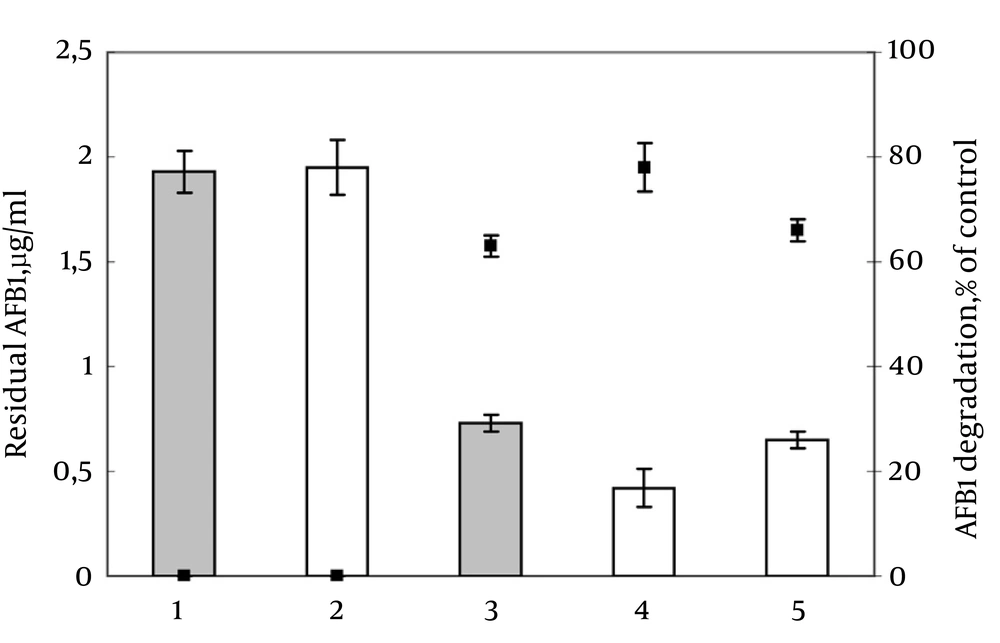
In all experiments, AFB1 degradation by PG41 was stable and reproducible. The ultrafilter-retained HMW metabolites accounted for 98.6% of the total AFB1-degrading activity; whereas the filtrates were inactive. Degradation of AFB1 positively correlated with the duration of incubation in CLF, the cell-free extract, or HMW metabolites (data not shown). AFB1–catabolizing activity in protein-enriched fractions of HMW metabolites, obtained from the CLF and cell-free extracts, showed that both HMW fractions degraded AFB1 (78% and 66%, respectively; Figure 1). The fraction from CLF was more active; hence, it was used in subsequent experiments.
AFB1-catabolizing activity of protein-enriched fractions of HMW metabolites from CLF was examined according to the effect of pH, temperature, and proteinase K treatment on the detoxifying activity (Table 1). The obtained results showed that the activity was pH-dependent, thermolabile, and significantly reduced by proteinase K treatment. In conclusion, these findings suggest that the studied extracellular metabolites are of protein origin and contribute to AFB1-degrading activity of PG41.
a pH value typical for culture liquid at the end of fermentation.
b Variants 2-4 were obtained by the acidification of initial P-CLF with 1N HCl.
5. Discussion
Use of AFB1-degrading enzymes to decontaminate products may overcome the disadvantages of using living microorganisms. Moreover, such enzymes can be manufactured under large-scale conditions. Yet, few attempts are made to determine the agents responsible for AFB1-degrading activity in bacteria and fungi.
Regarding bacteria, such activity was confirmed for metabolites of Stenotrophomonas maltophilia, Bacillus subtillis, Mycobacteriumfluoranthenivorans, and Rhodococcuserythropolis (9-11) including evidence of enzymes as an origin. All reported metabolites had a significant level of AFB1-degrading activity. The enzymatic AFB-degrading activity was also reported for metabolites of some basidiomycetes. While extracellular metabolites of Pleurotusostreatus and Peniophora sp. demonstrated low activity (35.9% and 25.8% within 72 hours, respectively), intracellular metabolites of Armillariatabescens completely degraded AFB1 within 30 minutes (9, 18). Furthermore, fungal laccase and manganese peroxidase isolated from Trametesversicolor (9) and Phanerochaetasordida (19) were reported to destroy 81.3% and 86% of AFB1 within 72 and 48 hours, respectively. Although there are several reports on sources of AFB1-degrading activity in bacteria and macromycetes, it seems that only one concerning micromycetes is available (12). In the latter study, 16 fungal cultures were assessed for AFB1-degrading activity. The most efficient strain (Phoma sp.) showed degradation activity by extracellular and intracellular metabolites (65% and 95%, respectively) similar to P. glomerata PG 41. At the same time, PG 41 showed better AFB1-detoxifying properties of its extracellular metabolites than that of the above study (63% degradation of added AFB1 (2 μg/mL) within 72 hours vs. 65% (0.5 μg/mL) within 120 hours (12)).
To date, basidiomycete A. tabescens is reported as the most promising AFB1-degrading activity . Also, some bacterial AFB1-degrading enzymes appear to act more rapidly than those of fungi. However, micromycetes are more considerable since they produce a wider variety of enzymes, which are often more stable than the ones of bacterial origin (20). Fungi are worthy enough to identify corresponding genes of AFB1-degrading enzymes. This knowledge could be used in genetic engineering of bacteria to overproduce degrading enzymes or in transgenic plants to limit pre-harvest AFB1 contamination. Culture liquid filtrate of P. glomerata PG41 isolated from a natural substrate inhabited by aflatoxigenic A. flavus, possesses significant AFB1-destroying activity. The revealed activity is associated mainly with a protein-enriched, high-molecular-weight fraction of extracellular metabolites and appears to be of enzymatic origin. Further isolation and identification of the enzyme(s) responsible for AFB1-degrading activity of P. glomerata is planned.