1. Background
Dental caries is well known as a biofilm-mediated disease; streptococci make up 60% to 90% of the supragingival plaque biomass in the first 24 hours of colonization (1, 2). Streptococcus mutans, as resident microflora and ubiquitous in the worldwide population, are biofilm-forming bacteria and are considered the primary etiologic agents of human dental caries (3, 4). Genetic diversity of S. mutans in plaques is reflected in their range of phenotypic adaptation to transmission, colonization, as well as different cariogenic traits among individuals (5, 6).
Initial acquisition of S. mutans happened in the window of infectivity in infancy by vertical transmission, mainly from mother (7). It might be suggested that horizontal transmission occurs in children, or adults (7, 8); however, exogenous microbes might be transient. Studies show that the colonization of indigenous flora is stable in teenagers and adults by genotype monitoring and follow-up inspection, and the transmission of exogenous microbes might be difficult to colonize permanently (9-11).
The interaction and communication should occur between the immune system and the microbiota: a driving force for evolution of the immune system is the need to accommodate diversity in a host microbiota; this in turn allows the host to accommodate environmental antigens and possibly self-antigens (12, 13). However, very little is known about the basic biology of naturally occurring host antibody in vivo to the persistence of the resident oral flora. The authors need to rethink how an immune system can aid in the colonization of indigenous strains and the exclusion of exogenous strains (14).
2. Objectives
Secretory IgA antibodies are the first line of mucosal defense against adherence and colonization by pathogens (15-17). The current study hypothesized that the secretory IgA should be the best defined effector component of the mucosal immune system to benefit from the permanent colonization of the natural flora, and the exclusion of alien flora. The current study aimed to identify the immunoblotting recognition profile of naturally occurring salivary IgA to indigenous and exogenous genotype strains of S. mutans using Chinese spousal pair model.
3. Patients and Methods
3.1. Study Population
Nine husband and wife pairs were randomly selected from volunteers to participate in the study in Wuhan city, China. Each subject (eighteen) signed a formal informed consent statement, and the entire study was approved by the medical ethics committee of Wuhan university. The inclusion criteria were that the couple had resided in the same household for at least five years before this study, had no chronic systemic diseases, did not take daily medicine and had not received any antibiotic treatment for two months. The dental examination for caries prevalence was carried out in fully equipped dental units using the world health organization (WHO) criteria for diagnosis and coding dental caries by one dentist. The dental caries experience i e, the sum of decayed, missing and filled teeth (DMFT) was recorded for each individual.
3.2. Saliva Samples
To minimize contamination with oral microbiota, 20 mL unstimulated submandibular/sublingual saliva was collected in the morning from each subject, using a modified collector as shown in Figure 1, and the flow rate was recorded. These samples were immediately transferred on ice and clarified by centrifugation at 6500 ×g for 30 minutes. The clarified saliva samples were stored at -70°C until use for the enzyme-linked immunosorbent assay (ELISA) and Western blot analysis. A sample of whole saliva was also collected from each subject and 10-fold serial dilutions were plated on mitis-salivarius-bacitracin agar (MSB) to estimate the level of mutans streptococci.
3.3. Identification of Indigenous Genotypes
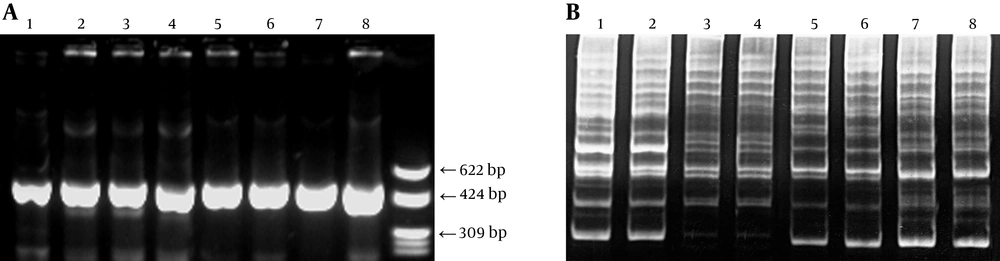
Clinical S. mutans strains in plaque were isolated, at least nine colonies were randomly selected based on their colony morphology on MSB plate from an individual, and the genomic DNA was extracted as reported previously (11). Serotypes of isolates were examined by biochemical tests including mannitol, sorbitol, raffinose, melibiose, arginine and mannitol containing bacitracin. Differentiation of S. mutans and S. sobrinus was confirmed by polymerase chain reaction (PCR) (18). Briefly, 1 μM/L primer; 4 μL (5’-ACTACACTTTCGGGTGGCTTGG and 5’-CAGTATAAGCGCCAGTTTCATC, 5’-GATAACTACCTGACAGCTGACT and 5’- AAGCTGCCTTAAGGTAATCACT); template DNA 2 μL, 5 U Taq DNA polymerase, 25 mM/L MgCl2 23 μL, dNTP 2 μL were diluted to a final volume of 50 µL. The amplification was programmed for 30 cycles (94°C for 30 seconds; 50°C for 1 minute; 72°C for 1 minute). Amplification products were identified by electrophoresis through 1.8% agarose gel (Takala Biotech, Japan) using the PCR-based molecular marker.
Genotypes of the isolates were identified using the arbitrarily primed PCR (AP-PCR) method. Briefly, 100 pmol primer OPA-02 (5’-TGCCGAGCTG, Takara Biotech, Japan), 2 µL DNA, 200 µM dNTP, 2.5 U Taq DNA polymerase, 7.0 mM MgCl2 were diluted to a final volume of 50 µL. The amplification was programmed for 35 cycles with the following conditions: 94°C for one minute; 36°C for two minutes; 72°C for two minutes. Amplicons were separated electrophoretically on 5% polyacrylamide gel using the PCR-based molecular marker.
The indigenous strain of S. mutans was determined by isolating it repeatedly in plaque samples from each subject, by monitoring genotype three times at three-month intervals.
3.4. Determination of Total Secretory IgA
Secretory IgA concentrations were measured by symmetrical capture ELISA. The microtiter plates (Nunc Roskilde, Denmark) were coated with rabbit anti-human α-chain antibodies overnight at 4°C, and then blocked with 5% bovine serum albumin (BSA) (Roche, Swiss) in Tris buffered saline (TBS). Serial 10-fold dilution of saliva samples were added in duplicate to plates and incubated overnight at 4°C. After washing, horse radish peroxidase (HRP)-conjugated goat α-chain specific anti-human IgA antibody (Sigma, USA, diluted in 0.05% Tween 20 in TBS) was added, and o-phenylenediamine was used for detection. The absorbance was recorded at 490 nm. A human milk sample with a known concentration of secretory IgA (Sigma, USA) was run simultaneously with the samples in order to generate a standard curve. Statistical analysis of the results was carried out between secretory IgA, salivary level of mutans streptococci and DMFT.
3.5. Western Blot Analysis
The indigenous isolates from subjects and the reference strains, including S. mutans GS-5 and S. mutans Ingbritt (C), were streaked on trypticase yeast-extract cystine agar medium (TYC, Difco, USA) and grown to mid-logarithmic stage in brain heart infusion (BHI) broth (pH 7.2). The bacteria were washed with phosphate buffered saline by centrifugation at 4°C and weighted; then, re-suspended and sonicated in an ice bath (Ultrasonic Processor VC130PB-1, 130W) for five minutes, and prepared by boiling in 2% sodium dodecyl sulfate (SDS), 5% 2-mercaptoethanol, 0.0012% bromophenol blue, 10% glycerol at 100°C for 15 minutes. Electrophoresis with 7.5% polyacrylamide gels was performed using protein molecular marker.
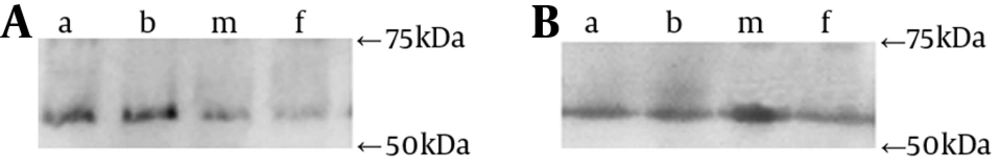
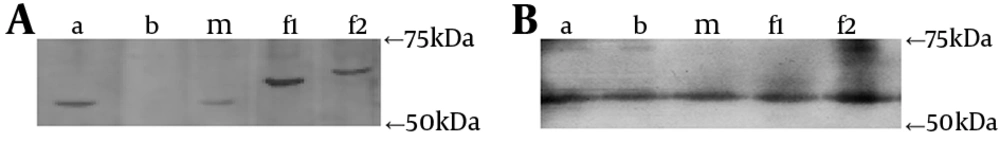
Salivary IgA antibody activity was detected using Western blot analysis. Each saliva sample was blotted to his or her personal indigenous strain, his or her spouse’s isolates (as exogenous strain donor), S. mutans GS-5 and S. mutans Ingbritt (C) as reference. Pilot screens were performed with different concentrations of the cell antigens to select the optimal concentration. Proteins were transferred from the gel onto polyvinylidene difluoride (PVDF) membranes (PALL-Gelman, USA). The membranes were rinsed and the nonspecific sites were blocked with 10% BSA. The PVDF membranes were washed in 0.5% BSA and TBS and incubated with undiluted saliva samples overnight. After washing in TBS, the membranes were incubated with 1: 3000 HRP-conjugated goat α-chain specific anti-human IgA antibody (Sigma, USA). The blot was incubated with Supersignal West Pico Substrate working solution (Pierce chemical company, USA) and exposed to film. The film was developed using appropriate developing solution and fixative for the type of film.
4. Results
4.1. Total Secretory IgA and Caries Prevalence
Table 1 presents the relationships among DMFT, length of marriage, salivary level of mutans streptococci (CFU/mL), flow rate of submandibular/sublingual secretion, the number of indigenous strains and the concentration of secretory IgA antibody in saliva. In nine couples, six individuals (females # 5, # 8, and males # 1, # 2, # 7, # 8) had no caries, with DMFT = 0. Female # 7 had high caries prevalence, with DMFT = 10 (D = 4, F = 6). The concentration of secretory IgA in saliva ranged from 33.3 to 90.7 µg/mL saliva. Salivary level of mutans streptococci and DMFT had no correlation with the concentration of secretory IgA in saliva (Spearman rank correlation: γs = -0.132, γ18, 0.1 = 0.401; P > 0.1).
An individual could harbor 1 - 2 indigenous S. mutans strains by genotype monitoring. A total of 20 unique indigenous genotypes were identified from 18 subjects, Figure 2.
| No. | Subject | Length of Marriage, Y | DMFT | Salivary Level of MS (CFU/mL) | Number of Indigenous Strain | Flow Rate of Submandibular/Sublingual Secretions (mL/min) | Secretory IgA (µg/mL) |
|---|---|---|---|---|---|---|---|
| 1 | Female 1 | 11 | 1 | 2.5 × 107 | 1 | 0.1 | 61.4 ± 1.1 |
| 2 | Male 1 | 0 | 0.5 × 107 | 1 | 0.16 | 53.2 ± 1.8 | |
| 3 | Female 2 | 21 | 9 | 6.7 × 106 | 1 | 0.2 | 66.5 ± 4.4 |
| 4 | Male 2 | 0 | 6.7 × 105 | 1 | 0.04 | 90.7 ± 2.7 | |
| 5 | Female 3 | 5 | 1 | 5.0 × 105 | 1 | 0.15 | 36.5 ± 2.7 |
| 6 | Male 3 | 4 | 3.2 × 107 | 1 | 0.1 | 67.4 ± 1.6 | |
| 7 | Female 4 | 6 | 1 | 5.0 × 105 | 1 | 0.1 | 55.4 ± 0.54 |
| 8 | Male 4 | 5 | 2.0 × 105 | 1 | 0.04 | 66.6 ± 2.1 | |
| 9 | Female 5 | 28 | 0 | 6.7 × 105 | 1 | 0.15 | 55.6 ± 3.1 |
| 10 | Male 5 | 4 | 1.0 × 107 | 2 | 0.03 | 33.3 ± 2.9 | |
| 11 | Female 6 | 17 | 7 | 1.0 × 105 | 1 | 0.14 | 60.1 ± 7.7 |
| 12 | Male 6 | 6 | 0.8 × 105 | 1 | 0.2 | 43.1 ± 9.2 | |
| 13 | Female 7 | 10 | 10 | 1.0 × 106 | 2 | 0.04 | 62.3 ± 6.3 |
| 14 | Male 7 | 0 | 6.6 × 105 | 1 | 0.1 | 57.6 ± 9.9 | |
| 15 | Female 8 | 36 | 0 | 2.6 × 105 | 1 | 0.13 | 71.5 ± 2.7 |
| 16 | Male 8 | 0 | 4.0 × 105 | 1 | 0.1 | 75.9 ± 0.7 | |
| 17 | Female 9 | 6 | 6 | 5.9 × 105 | 1 | 0.04 | 56.5 ± 5.0 |
| 18 | Male 9 | 1 | 4.0 × 105 | 1 | 0.167 | 65.5 ± 3.0 |
Abbreviations: DMFT, decayed, missing and filled teeth; MS, mutans streptococci.
aNumber of indigenous strain: the number of genotype which colonized stable by AP-PCR monitoring three times at three-month intervals.
4.2. Individual secretory IgA Reaction to Different Streptococcus mutans Genotypes
Almost all subjects had similar individual immunoblotting profiles of secretory IgA to different S. mutans genotypes, including his own indigenous strain, the spouse’s genotype, S. mutans GS-5 and Ingbritt (C), with the detectable blot bands and the most intensity discrimination as natural higher-affinity in the domain of 55 - 75 kDa (Figure 3). Using the spousal pair model, only the male # 7 with no decayed caries (D = 0), had different particular patterns of two genotypes comparing those of his wife, with special blotting bands in the domain of 55 - 75 kDa (Figure 4). These results indicated that certain epitopes, in the domain of 55 - 75 kDa, might be important antigens recognized by natural salivary IgA.
5. Discussion
In the current study, married couples were recruited, and the indigenous strains were characterized by genotype monitoring. During the couple’s daily life, frequent and close contact lead to frequent inoculation of each other’s S. mutans strain. Each person is a receptor to one’s own indigenous genotype, as well as a donor of exogenous genotypes to one’s spouse. This spousal ± pair model is of specific value in investigating the different responses of host nature immune system to both endogenous and exogenous strains.
The study also used a modified plastic collector to ensure that submandibular/sublingual saliva had not been in contact with the subject’s dental plaque and to avoid exposure to oral bacteria; therefore, the saliva testing reflects the actual immune responses of the subjects in vivo. These were different forms of what Widerstrom et al. (19, 20) had done by one sampling.
Authors’ previous studies involving genotype monitoring of S. mutans showed that the colonization of indigenous genotypes could be stable in adults, despite frequent and close contact between husband and wife, and the transmission of exogenous microbes might be transient due to the difficulty to colonize permanently (11). Another longitudinal study in teenage subjects undergoing orthodontic treatment confirmed and extended the findings of others regarding the genotypic stability of the S. mutans. This population of subjects is somewhat unique since the wearing of orthodontic appliances should increase plaque accumulation, elevate mutans streptococci colonization, enhance the susceptibility to white ± spot formation, and induce decreased pH (9). Then another study on patients with nasopharyngeal carcinoma also confirmed that most genotypes of S. mutans were persistent after radiotherapy, thought ionizing radiation in the head and neck region can destroy the function of salivary glands and result in a reduction of salivary flow (10). Nevertheless, the mechanisms underlying the fact that the indigenous flora was accepted and exogenous flora was expelled by the host is unclear, and results are ambiguous as to whether the secretory immunity system is generally effective in preventing colonization and transmission of microbes.
The indigenous microflora includes a relatively large number of genetic variants (21). It was also demonstrated that the genetic diversity of S. mutans resulted in phenotype diversity and protein production variety (22). The relationship between transmission and colonization of S. mutans, dental caries and natural immunity are complicated by the bacterial genetic diversity among population.
Secretory IgA is the best defined effector component of the mucosal immune system to defend against infectious agents and other harmful substances (17). It is reasonable to assume that secretory IgA would benefit from homeostasis of indigenous strains and expel exogenous strains. Therefore, it was thought different Western ± blotting patterns among S. mutans genotypes and saliva might indicate differences in colonization, which can be a reasonable natural selection of host immune system and could be related to secretory IgA modulation of colonization.
Nonetheless, the evidence of this pilot investigation and monitoring would point to the conclusion that secretory IgA might have no direct correlation with the colonization of indigenous flora and rejection of exogenous strains in adults. From the results of this study, almost all subjects had similar immunoblotting profiles of secretory IgA to isolate and refer to S. mutans strains. Total salivary IgA concentrations were determined in order to provide a standardized basis to compare different samples, and flow rates were also recorded. High salivary flow rates and high concentration of antibodies benefit from the clearance of exogenous microbes and interfere with adherence (23, 24). Other factors such as individual’s heath, doing exercise and diet might affect the level of IgA (25-27). However, neither has definitive role on the homeostasis of oral micro ± ecosystem, which is derived from of various factors in ecological systems. It was not a surprise of this pilot investigation that the concentration of secretory IgA in saliva did not correlate with salivary level of mutans streptococci and DMFT using ELISA test.
First, it might be mostly due to the similar immunogenicity of different strains or immune cross ± reaction of species belong to the same species (14). Additionally, the Western blotting might focus only on common antigens. If more commonality of genotypes were classified among population, the comparison of saliva might be associated with differences of the host. Unfortunately, the better discrimination ability of methods are well ± accepted and used, such as restriction endonuclease analysis, ribotyping, AP ± PCR, etc., more excessive diversity of genotype would be observed (8). Twenty genotypes from 18 subjects were found in this investigation.
Second, specific antigens of S. mutans are usually less than 1% of the total IgA level in saliva. Also it seems that accepting or expelling of some kind of genotype strains is not only a direct result of the secretory IgA antibody activity. It is suggested to limit the induction of host immune response to common antigens; therefore, likely not involve in adherence among the mechanisms that commensal streptococci employ to persist in the oral cavity.
Third, the endogenous strain has adapted itself to an environment of stress and has become a commensal strain to colonize in a state of homeostasis (14). Since the ecological niche is already filled with the indigenous strain, it might be difficult for an exogenous S. mutans strain to take the place of the indigenous one, even though it may be a special genotype with more immunogenic or cariogenic traits.
The reaction of secretory IgA with antigens in the domain of 55 - 75 kDa, might be worth a detailed study. These antigens may be glucan ± binding proteins (GbpB 59 kDa and GbpC 63.5 kDa) or fragments of other surface antigens according to their molecular weight (28, 29). The latest study showed that distribution of putative virulence genes in S. mutans strains does not correlate with that of caries experience (22). It should be explored and clarified since some studies discussed and demonstrated strong immunogenicity or immunoreactivity of some proteins in experimental models and laboratory investigations.
In conclusion, the current study indicated that naturally induced salivary IgA antibodies against S. mutans may be present in all subjects, but its concentration and immunoblotting profile might have no direct correlation with the colonization of indigenous flora and rejection of exogenous strains in most adults. Despite the fact that S. mutans have genotypic and phenotypic diversity as principle bacterial pathogen, it might have no significant impact on the natural immune system of the host with a mature and stable ecosystem in oral cavity. The colonization and transmission of S. mutans should be in the light of coevolved microecosystem as a whole, but not caused by one factor alone.