1. Background
The highly neurotropic Rabies virus (RABV) is the most significant human pathogen of the Rhabdoviridae family and is a prototype member of the Lyssavirus genus. RABV is the causative agent of rabies, which remains a global public problem with an estimated human death toll of more than 55000 annually (1). The RNA genome (~ 12 kb) of RABV encodes five viral proteins, including Nucleoprotein (N), Glycoprotein (G), Phosphoprotein (P), Matrix protein (M), and RNA-dependent RNA polymerase (L). It has been shown that the transcription and replication of RABV take place within Negri-Body-Like (NBL) structures, which are inclusion bodies formed in the process of RABV infection (2). The NBL structures of RABV-infected cells are composed of the viral nucleocapsid, viral genome, and a series of cellular proteins (3).
Prefoldin binds specifically to cytosolic chaperonins (e.g. chaperonin TRiC/CCT) and transfers the target proteins to it. Prefoldin binds to the nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins. Prefoldin subunit 1 (PFDN1) was shown to be one of the differential proteins in the Vaccinia virus IHD-W infected HEK 293 cells (4). The hepatitis C virus (HCV) F protein was found to interact with cellular protein PFDN2 through the use of the yeast two-hybrid system and perturb the tubulin cytoskeleton organization (5). PFDN1 and PFDN2 are two of the upregulated and downregulated differentially expressed proteins between a hepatitis B virus (HBV)-producing cell line HepG2.2.15 and its parental cell line HepG2, respectively.
Viruses are fully reliant on the successful recruitment of host cellular factors in their viral life cycle, and viruses usually seize control of the host signaling pathways and cellular translation factors, which regulate their activity (6). Host protein Hsp70 is present in both purified RABV virions and in nucleocapsids purified from infected cells, and it also has been shown to interact with the N protein of RABV (7). Dynein light chain 8 (DLC8) has been reported to bind to the P protein of RABV and promote efficient viral transcription (8-10). The M of RABV plays a role in inhibiting translation in virus-infected cells through an interaction with the eukaryotic translation initiation factor 3, subunit H (eIF3h) (11). Recently, we found out that the subcellular distribution of host proteins CCTγ (3) and CCTα (12) was altered in RABV-infected cells and that it played a role in RABV infection. However, we detected no direct interaction between CCTγ or CCTα and the viral proteins N or P. We are, therefore, still in the dark as to whether the upstream of chaperonin CCT, Prefoldin, has a similar subcellular redistribution in RABV-infected Neuro-2a (N2a) cells.
2. Objectives
Besides viral proteins, host cellular factors play a key role in the replication of various viruses. The aim of the present study was to investigate the subcellular distribution and transcription of the chaperone PFDN1 in RABV-infected N2a cells.
3. Materials and Methods
3.1. Cell Culture, Viruses, Antibodies, and Reagents
Mouse N2a cells were grown at 37°C in a humidified CO2 (5%) incubator with completed Dulbecco’s modified eagle’s medium (DMEM) (Invitrogen, CA, USA) supplemented with 10% (V/V) fresh newborn calf serum (Sijiqing, Hangzhou, China). The RABV HEP-Flury strain was propagated in N2a cell monolayers. Rabbit polyclonal Antibody (pAb) against PFDN1 was purchased from ProteinTech Group Inc. (Chicago, IL, USA), Rabbit monoclonal Antibody (mAb) against DLC8 was purchased from Epitomics Inc. (Burlingame, CA, USA), and 4', 6-diamidino-2-phenylindole (DAPI) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse mAb anti-N protein of RABV was produced as described previously (3, 13).
3.2. Transfection
One day before transfection, the cells were plated at a density of 104 per well containing a 10% fetal calf serum DMEM without antibiotics. On the day of transfection, the culture should have 80% confluency. The pCI-neo-N and pCI-neo-P vectors were cotransfected or monotransfected into the N2a cells using Lipofectamine 2000 (Invitrogen, USA). Forty-eight hours later, the cells were fixed with ice-cold acetone-methanol (1/1) for subsequent use in immunofluorescence or directly extracted RNA for real-time quantitative polymerase chain reaction (qPCR).
3.3. Co-Focal Microscopy Assay
The mouse N2a cells were grown in a 96-well plate to 80% - 90% confluence before being infected with RABV at a multiplicity of infection (MOI) of 1. Cultured for 48 hours, the cells were washed with phosphate-buffered saline, fixed with pre-chilled acetone-methanol (1/1) at 20°C for 20 min, and air dried. After having been blocked by 5% skimmed milk, the cells were incubated with mouse mAb against the N gene of RABV and rabbit pAb against PFDN1 or rabbit mAb against DLC8 for 2 hours. The nuclei of the cells were stained with DAPI for immunofluorescence assay.
3.4. Real-Time Quantitative Polymerase Chain Reaction
To investigate whether the transcription of PFDN1 was upregulated or downregulated in the RABV-infected and the N and/or P gene of the RABV-transfected N2a cells, the total RNA of the N2a cells was infected with RABV or transfected with the N or P gene for 48 hours. Also, cDNA synthesis was performed using oligo (dT) primers (100 ng) with a RevertAid first strand cDNA synthesis kit (Fermentas, Thermo Scientific), and real-time qPCR was performed as described previously (3). Briefly, it was carried out on the 7500 real-time PCR system with 200 ng of cDNA template, 1 × SYBR premix Ex taq (perfect real time, TaKaRa, Dalian, China), and 200 nM of each primer in the volume of 20 μL. Specific primers (Table 1) were designed and synthesized by Shanghai Shenggong (Shanghai, China) to simultaneously amplify various target genes. The amplification conditions were as follows: 95°C for 10 min; followed by 40 cycles of 95°C for 10 seconds, 60°C for 30 seconds, and 72°C for 10 seconds. The melting curve was recorded after the PCR was run. And relative expression was calculated by the comparative ∆∆Ct method.
| Gene Symbol | Forward Primers (5’ → 3’) | Reverse Primers (5’ → 3’) |
|---|---|---|
| GAPDH | TCAACAGCAACTCCCACTCTTCCA | ACCCTGTTGCTGTAGCCGTATTCA |
| DLC8 | TGTCGGAAGAGATGCAACAGGACT | ATGGGCCGCAATATCCTTCTCGAT |
| PFDN1 | AGGTCCCAACTGAATGTCTGCCAA | AGGCTTGGACAGCTTTCTCCATCT |
a Abbreviations: Dynein light chain 8; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; DLC8, PFDN1, Prefoldin subunit 1.
4. Results
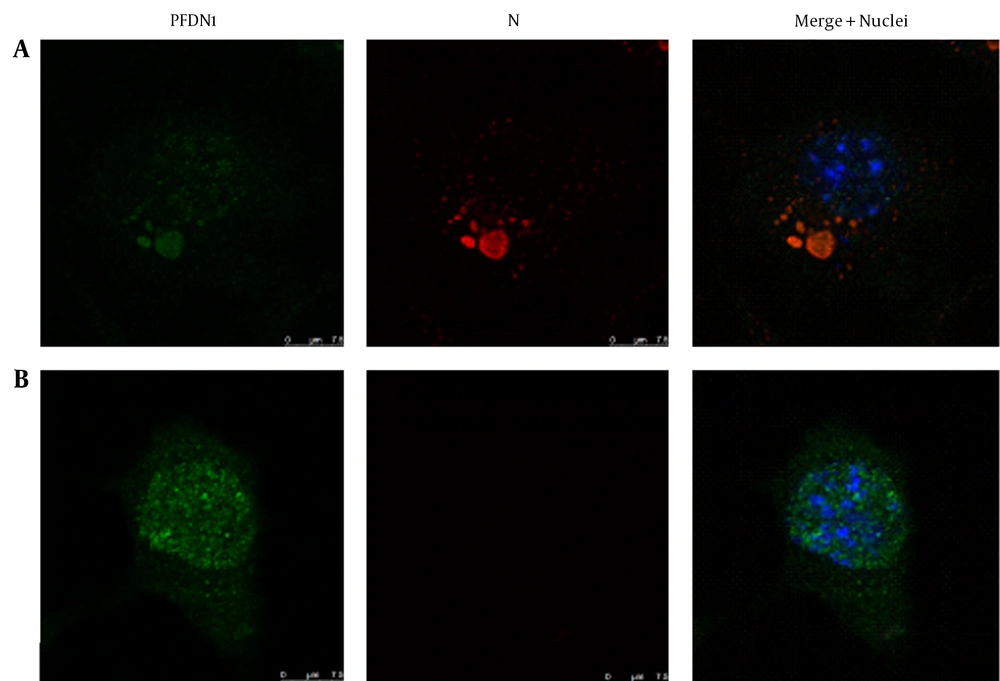
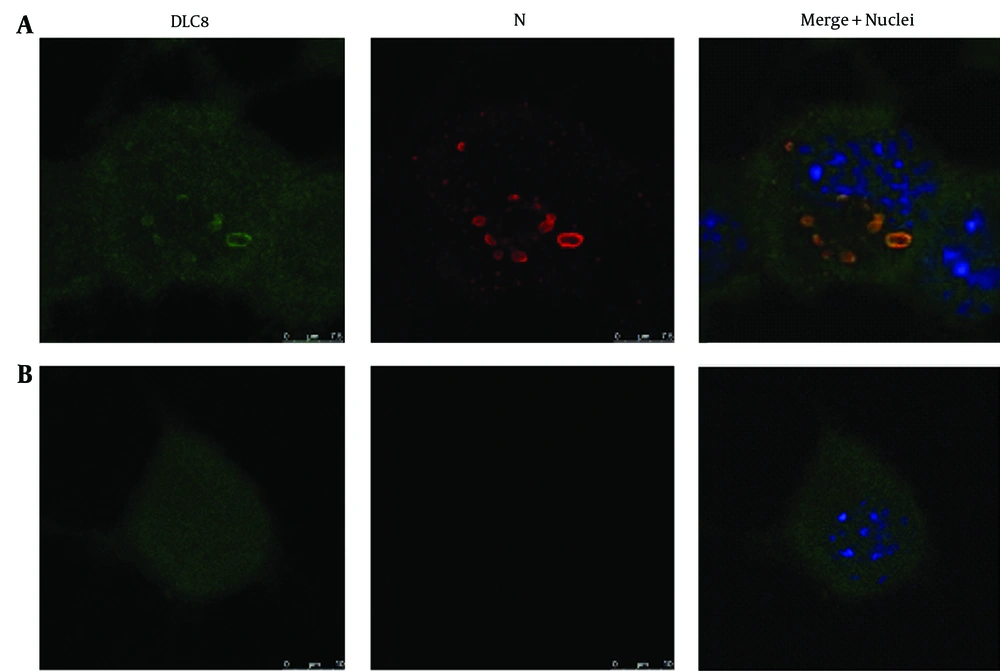
4.1. Subcellular Redistribution of Prefoldin Subunit 1 and Dynein Light Chain 8 in the Rabies Virus-Infected Neuro-2a Cells
Given the similarity between chaperone complex Prefoldin and chaperonin complex TRiC/CCT, as well as their cooperation in protein folding, the question is whether an alteration of Prefoldin distribution occurred in the RABV-infected N2a cells. DLC8 has been reported previously by several research teams to be a partner of the P gene of RABV and used as a positive control. Immunostaining was conducted using mAb to the N protein of RABV, followed by Tetramethylrhodamine (TRITC)-conjugated IgG (red fluorescence) and counterstaining using rabbit antibodies to PFDN1 and DLC8, followed by FITC-conjugated IgG (green fluorescence). The cell nuclei were stained with DAPI (blue fluorescence). The triple-stained cells were observed by Leica TCS SP5 laser confocal microscopy. The relevance of PFDN1 and DLC8 to the localization of the N protein of RABV was analyzed by confocal microscopy (Figure 1 A and 2 B and Figure 2 A and 2 B). In the RABV-infected N2a cells, the neuroblastomas (NBs) containing the viral N gene in the cytoplasm was colocalized with intracellular protein PFDN1 (Figure 1 A) and DLC8 (Figure 2 A). In the mock-infected N2a cells, PFDN1 was distributed both in the nucleus and cytoplasm; however, PFDN1 was mainly distributed in the cytoplasm of the N2a cells (Figure 1 B), while DLC8 was homogeneously distributed in the cytoplasm and nucleus of the N2a cells (Figure 2 B).
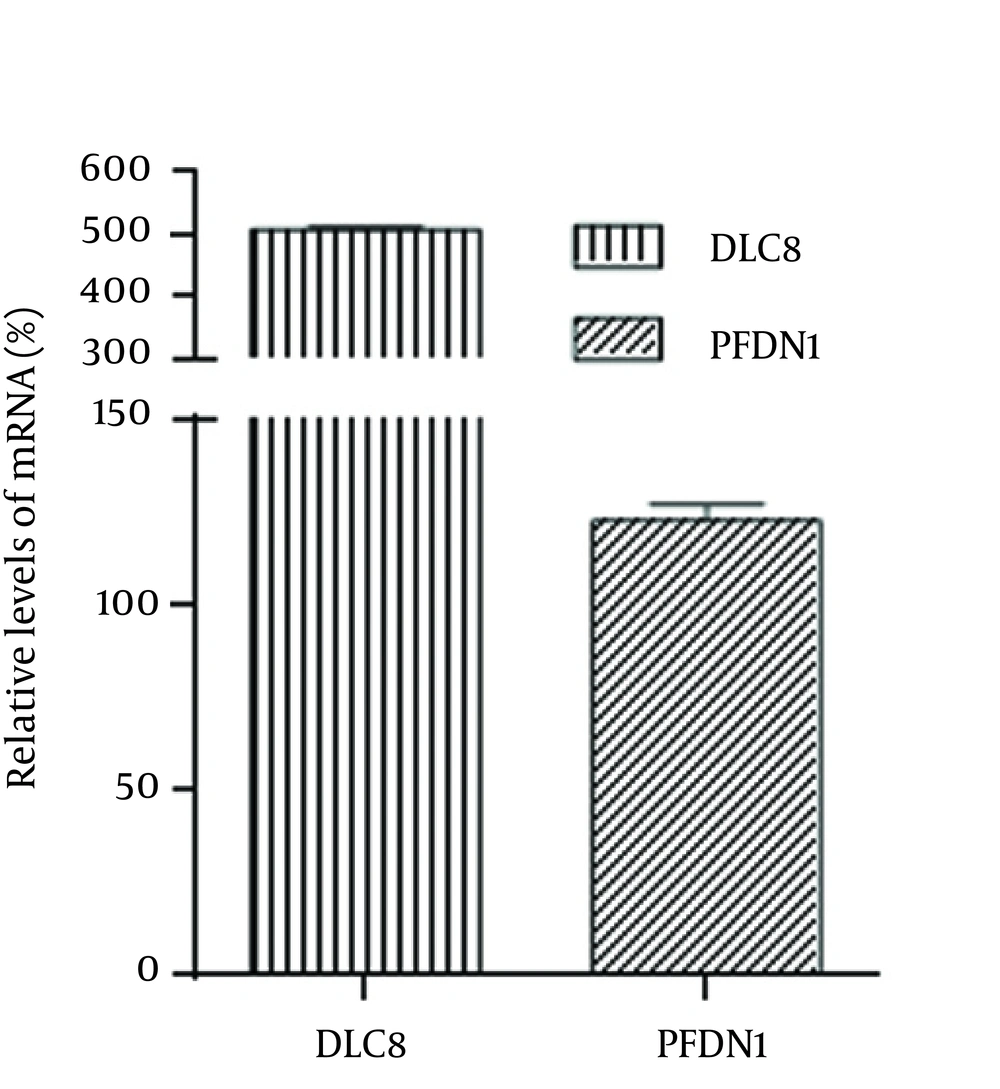
4.2. Transcriptional Alterations of Prefoldin Subunit 1 and Dynein Light Chain during Rabies virus Infection
To determine the change of PFDN1 and DLC8 in transcriptional levels, the genes encoding PFDN1 and DLC8 were quantified by comparative real-time PCR (Figure 3) using primers shown in Table 1. The mRNA transcript of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control housekeeping gene. Both of the two proteins are upregulated in varying degrees. The transcript abundance of PFDN1 in the RABV-infected N2a cells was raised 1.23 times as against the abundance of the mock-infected N2a cells, whereas the transcript abundance of DLC8 in the RABV-infected N2a cells was increased fivefold as compared with the abundance of DLC8 in the mock-infected N2a cells.
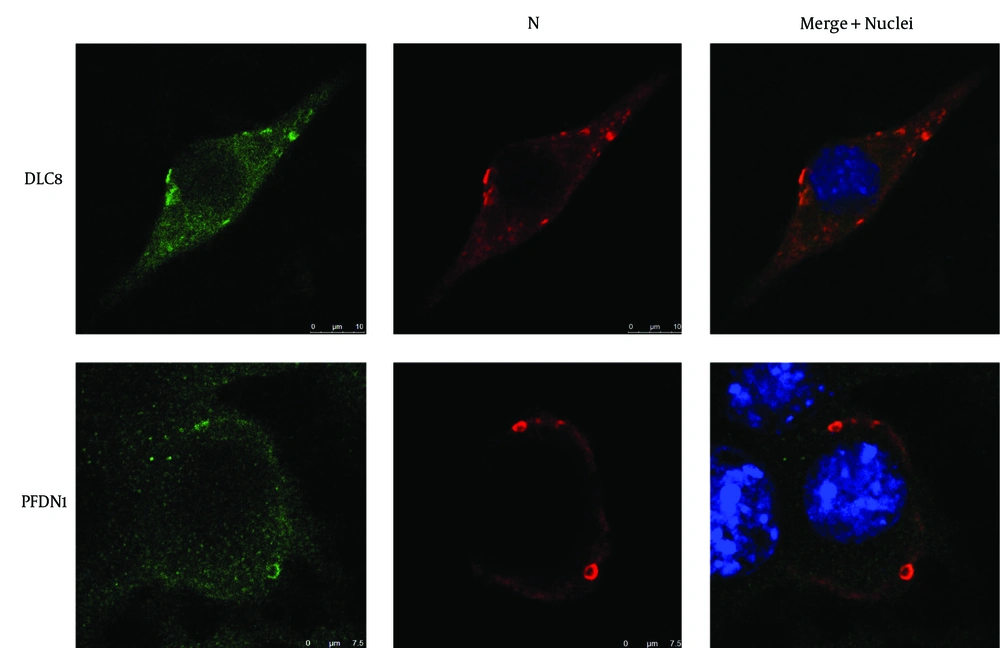
4.3. Co-Localization of Prefoldin Subunit 1 and Dynein Light Chain 8 with Negri-Body-Like Structures in the Plasmids Nucleoprotein- and Phosphoprotein-Cotransfected Cells
We have previously demonstrated the re-localization of CCTγ in cotransfected N and P genes of RABV in N2a cells (3). To determine whether proteins PFDN1 and DLC8 were involved in the formation of NBL structures, we used immunofluorescence to check the subcellular distribution of the PFDN1, DLC8, and N gene of RABV in pCI-neo-N and pCI-neo-P cotransfected cells. In the N and P cotransfected N2a cells, the NBL structures were formed, which induced the co-localization of PFDN1 and DLC8 with the N protein of the NBL structures (Figure 4). These results imply that PFDN1 and DLC8 are directly or indirectly involved in the formation of Negri bodies at early stages.
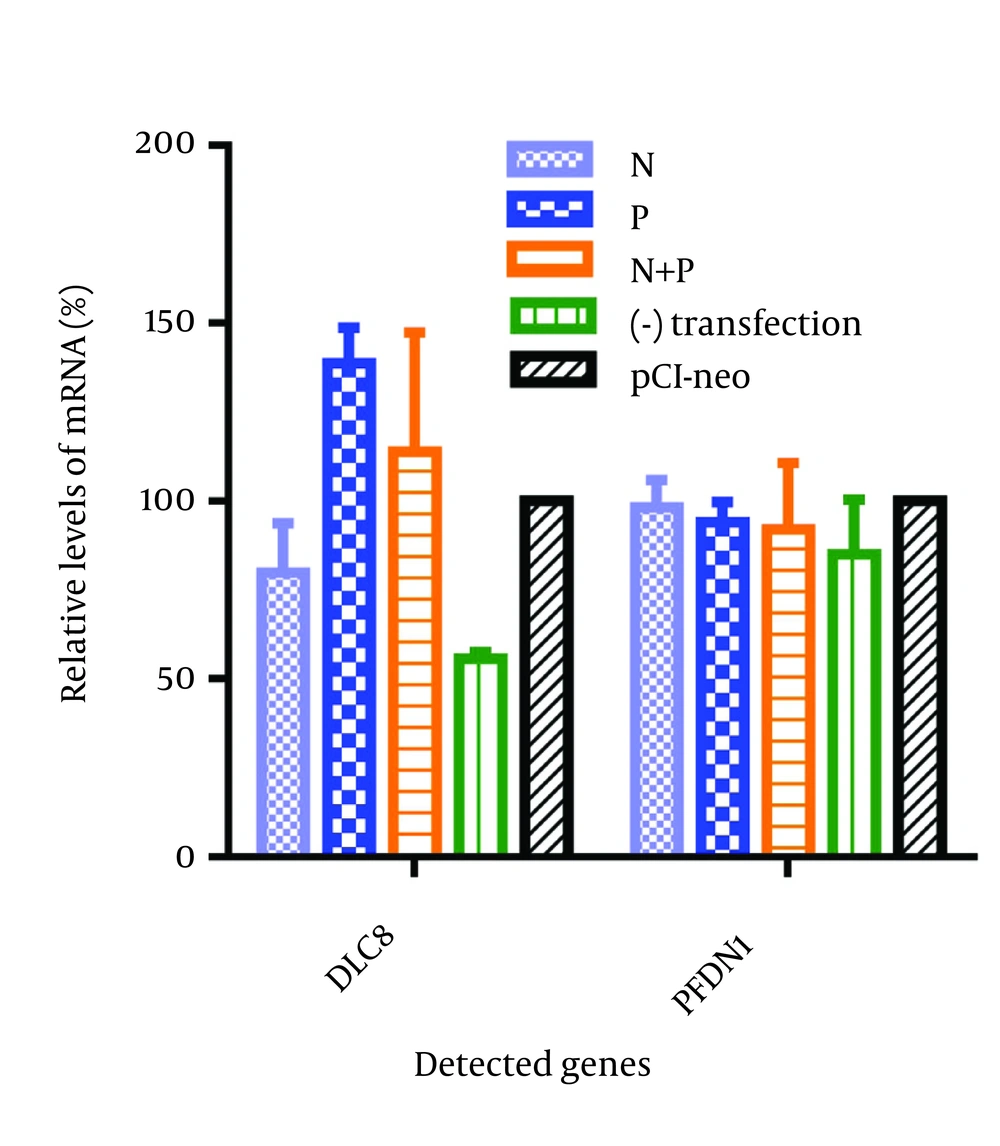
4.4. Transcriptional Alterations of Prefoldin Subunit 1 and Dynein Light Chain 8 in the Plasmid-Transfected Cells
We next investigated whether the transfection of the N and P genes of RABV individually or their cotransfection could induce the upregulation of the PFDN1 and DLC8 in the N2a cells. The mRNA transcript of PFDN1 and DLC8 was quantified by comparative qPCR. The gene transcriptional levels of the PFDN1 and DLC8 in the pCI-neo-transfected N2a cells were normalized to 100%. Compared with the pCI-neo-transfected cells, both of the cells monotransfected with plasmid pCI-neo-P and cotransfected with plasmid pCI-neo-N were able to induce the upregulation of DLC8 by 38.78% and 13.98%, respectively (Figure 5). Nevertheless, the transfection had no significant effect on the transcript of PFDN1 (Figure 5). Additionally, compared with the non-transfected cells, our results showed that the transfection itself was able to induce the host genes’ upregulation at vary levels, as is shown in Figure 5.
5. Discussion
In this study, we used co-focal laser microscopy and real-time qPCR to investigate the alterations of subcellular distribution and transcription of host cellular factors of PFDN1 and DLC8 in RABV-infected and plasmids containing the N and P genes of RABV-transfected N2a cells. In two previous studies (3, 12), we determined that the chaperonins CCTγ and CCTα were re-localized to the NBL structures of RABV-infected and transfected cells, which implies that the chaperone may play a role in the life cycle of RABV. However, we observed no direct interaction between the host protein of CCTα and CCTγ and the N or P protein of RABV through co-immunoprecipitation. This finding seems to be in consequence of an indirect interaction between the up or downstream of TRiC/CCT and viral proteins. It is well accepted that PFDN1 plays an important role in the correct folding of other proteins. It binds specifically to cytosolic chaperonins and transfers the target proteins. The results of the present study were exactly as we had predicted: PFDN1 was redistributed to the NBs in the RABV-infected N2a cells. Although we do not have experimental evidence to explain this phenomenon, given the accumulation of PFDN1 in the cytoplasm NBs observed upon infection, it is tempting to suggest that the virus may have triggered the stress-response protein of the cell against unfolded proteins.
We then asked whether the relocalization of PFDN1 and DLC8 to the cytoplasm NBs structures in the RABV-infected cells was due to the presence of one or two viral proteins of RABV. It should be mentioned that similar immunofluorescence tests were carried out on RABV infection and the plasmids of the RABV-transfected N2a cells. Furthermore, the co-expression of the N and P genes of RABV is a key factor for inducing the redistribution of PFDN1 and DLC8. All viruses are dependent on the host cell machinery for their replication and as such modify a series of cellular pathways (14). Remarkably, the results of the current study revealed an increase in the transcript abundance of PFDN1 and DLC8 in the RABV-infected cells. In order to find the viral factor that induced the upregulation of PFDN1 and DLC8, we tested the transcript of the host genes in the N and P genes of the RABV-transfected N2a cells. The results were interesting inasmuch as the co-expression of the N and P genes of RABV was able to induce the upregulation of DLC8, while PFDN1 was not affected significantly. A possible explanation is that probably the minor upregulation of PFDN1 in the RABV-infected cells was induced by the presence of the other proteins of RABV such as G, M, and L. The protein complex of Prefoldin was also found to be upregulated in the brains of patients with Alzheimer's disease (15).
Prefoldin is a jellyfish-shaped heterohexameric co-chaperone of group II chaperonins and is known to assist in capturing protein-folding intermediates and transferring them to group II chaperonins for the completion of folding (16). The manner in which Prefoldin cooperates with the chaperonin and chaperone and interacts with its substrates is poorly understood. Chaperone HSP70 has numerous co-chaperones that support its chaperone function in protein folding. Prefoldin is also one important group of co-chaperones of HSP70 (17). Previously published data by our group showed that the subcellular redistribution of CCTγ and CCTα played a role in RABV replication (3, 12). In addition, Lahaye et al. (7) reported that HSP70 interacts with the N protein of RABV and suggested that a variety of host chaperonins and chaperones are involved in the replication of RABV. Indeed, Prefoldin is an intermediary factor between HSP70 and TRiC/CCT (17). This could explain why we failed to confirm the interactions between the viral proteins of RABV and CCT.
We speculate that the recruitment of PFDN1 to NBs could also potentially assist in the folding of a variety of viral proteins or host proteins from their nascent polypeptides during the replication of the virus. Our findings provided evidence that upon RABV infection, another chaperone, PFDN1, is redistributed to the cytoplasm NBs, where the interacting partners of the viral replication complexes (i.e. viral RNA N, P, and L) localize.