1. Background
Candida species are the most common cause of opportunistic fungal infections and Candida albicans is the most predominant fungal opportunistic pathogen (1). However the relative prevalence and incidence of the Candida species depends on the geographical location, patient population, and clinical settings (2-4). In recent years, an increase is observed in invasive mycosis, particularly Candida infections (5-9). The increase is accompanied by a shift towards a higher proportion of species other than C. albicans, which are frequently resistant to fluconazole. C. krusei is one of the species that inherited resistance to fluconazole and is mainly isolated from immunocompromised patients such as patients with hematologic malignancies (10, 11).
Widespread use of fluconazole to prevent fungal infections in patients with human immunodeficiency virus infection contributes to a significant increase in C. krusei infection, especially due to the high incidence of resistance to this drug (12). Many of classes of anti-fungal drugs are currently available to treat fungal infections. These classes include polyene, azoles, allymines and echinocandins (13). The azole compounds, especially fluconazole are the most frequently used anti-fungal agents (14). Although the azole drugs have good pharmacokinetic properties and low toxicity, they are fungistatic and inhibit only the growth of the fungal cells and do not kill them. A critical problem is the resistance of fungal pathogens to antifungal drugs especially azole drugs. Currently, C. krusei is considered as a resistant to the azole drugs (15). The mortality rate due to fungal infections remains very high because of the limitations of effective available drugs (16).
The production and development of an effective fungicidal drug requires the identification of an essential fungal protein as drug target (17). Aconitate hydratase (ACO) is a mitochondrial protein that plays a vital role in tricarboxylic acid (TCA) cycle and thus production of energy within the cell (18). There are two forms of the protein aconitase (aconitate hydratase) in eukaryotes such as fungi: a mitochondrial and a cytoplasmic form. Mitochondrial aconitase convert citrate to isocitrate via a cis-aconitate intermediate in the TCA cycle. The cytoplasmic aconitase protein, in addition to enzymatic activity, is also an iron regulatory protein (IRP) (19). Targeting the fungal aconitase selectively would lead to the rapid death of fungal cells with little or no toxicity to the human host. It is necessary to obtain selective inhibition to determine any amino acid residue differences between human and fungal aconitases. These amino acids could then be used as a target to selectively bind and inhibit the function of fungal aconitase, thus produce the basis to innovate a new antifungal drug class.
Several studies commonly indicated that C. krusei virulence was less than that of C. albicans in terms of its adherence to epithelial cells and prosthetic surfaces, proteolytic activity and production of phospholipases. Furthermore, it seems that structural and metabolic features of C. krusei are significantly different from those of other medically common Candida spp. and display different behavioral patterns to host defenses, adding credence to the belief that it should be reassigned taxonomically (12, 20, 21). rapid amplification of cDNA ends (RACE) is a method to amplify cDNA fragment from an mRNA template with a known internal site and unknown sequences at either the 3’ or 5’-end of the mRNA (22). This procedure is defined by other researchers as one-sided PCR (23) or anchored PCR (24).
2. Objectives
The current study employed 5’ RACE method and degenerate PCR to partial lysequence the C. krusei aconitase gene. The current study aimed to identify C. krusei taxonomic location based on ACO gene sequence, determine any structural differences between C. krusei, common pathogenic Candida spp. and human aconitase sequences, and produce the basis to create a new antifungal drug to block the C. krusei aconitase function.
3. Materials and Methods
3.1. Strains
The current study used the standard strain of C. krusei (ATCC: 6258). The yeast was cultured on yeast extract (5g/L, Baltimore Biological Laboratory, USA)/ peptone (10g/L, Merck, Germany)/dextrose (20g/L, Merck, Germany) agar (20g/L, Merck, Germany) (YEPD) plate, incubated at 37°C for 48 hours, and then transferred into yeast extract peptone dextrose broth at 37°C for 24 hours.
3.2. 5’ Rapid Amplification of cDNA Ends (5’ RACE) Method
The used 5’ RACE method in this study was introduced by Xiang Zong Shi and Jarvis to identify GC rich sequences (25).
3.2.1. RNA Extraction
Total RNA was extracted from the exponential phase of yeast using the RNeasy Protect Mini Kit (Qiagen, Germany). For mechanical disruption, the yeast cells were sonicated (Hieschler, Germany; cycle 0.6; Amplitude 70%) with acid-washed glass beads (0.4 - 0.5 mm diameter). The quantity and quality of the extracted RNA was measured using the Nanodrop 1000 spectrophotometer (Thermo Scientific, USA).
3.2.2. cDNA Synthesis
For complementary DNA (cDNA) synthesis in a total volume of 20 μL, 2 μg of total RNA from organism was heated at 65°C for 7 minutes, followed by cooling on ice. A master mixture contained 4 μL of 5× reverse transcriptase (RT) buffer (Fermentas, Canada); 1 μL of gene-specific primer (5 ACO1R1; Table 1) (10 pmol/μL); dNTP Mix, 1 μL (10 mM); Ribolock, 1 μL(20 U) (Fermentas, Canada); and diethylpyrocarbonate (DEPC) treated water. Four hundred units (2 μL) of Moloney murine leukemia virus (M-MuLV) reverse transcriptase enzyme (Fermentas, Canada) were added. The RT temperature was set to 42°C for 1 hour and 70°C for 7 minutes. Subsequently, the reaction was treated with an equal volume of 0.6 N NaOH for 20 minutes at 65°C to degrade the mRNA and then the cDNA was precipitated by adjusting the solution to 0.5 M NaCl and adding absolute ethanol (2.5 times the total volume). After incubation at −80°C for 2 hours, the cDNA was separated using centrifugation for 30 minutes at 13,000 × g, washed with 70% ethanol, and dissolved in 10 μL of a commercial ligation-adaptor provided as follows:
| Name | Tm | Sequence (5’ to 3’) | Reference |
|---|---|---|---|
| UPM Long | 77°C | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | (23) |
| UPM Short | 53°C | CTAATACGACTCACTATAGGGC | (23) |
| Sense Adaptor (SA) | 71°C | AAGCAGTGGTATCAACGCAGAGTGNNNNN | (23) |
| Antisense Adaptor (ASA) | 62°C | p-ACTCTGCGTTGATACCACTGCTT (5’-phosphorylated) | (23) |
| 5ACO1R1 | 57°C | GTTCAAGTCAATGGCTCTGG | In this study |
| 5ACO1R2 | 52°C | AGTGGACAGTAGATGGAGTGG | In this study |
| 5ACO1R3 | 58°C | TGGGCGGTAGCATCTTGAC | In this study |
| 3ACO1F | 54°C | ACCCAAGATATTGAAAGAGG | In this study |
| 3ACO1R | 58°C | AATCTTGGTTCCAAAGCAGC | In this study |
| M13 F | 54°C | GTAAAACGACGGCCAG | Universal |
| M13 R | 56°C | CAGGAAACAGCTATGAC | Universal |
3.2.3. Adaptor Ligation
The ligation adaptor was provided by mixing two commercial oligonucleotides, named sense adaptor and antisense adaptor (Table 1), in an annealing buffer comprised of 10 mM Tris-HCl (pH. 8.0) and 50 mM NaCl. The mixture was placed in a boiling water bath, which was then allowed to slowly cool to room temperature, and the resulting adaptor was added to the first-strand cDNA, as explained above. Subsequently, a ligation reaction was done in a total volume of 10 μL, using 3 μL of the cDNA-adaptor mixture, 10X T4 DNA ligase buffer, 1.0 μL; and T4 DNA ligase, 1.0 μL. This mixture was placed overnight at room temperature and then the adaptor-ligated cDNA was diluted fivefold with ddH2O and used as the template for subsequent PCRs.
3.2.4. PCR amplification
Primary PCR mixtures included 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon), 2.0 μL of the diluted adaptor ligated first-strand cDNA described above, 2 μL of a gene-specific primer (5ACO1R2; Table 1), 2 μL of a primer complementary to the 3’ end of the adaptor sequence (UPM long; Table 1) and 6.5 μL of ddH2O in a total volume of 25 μL. Touchdown cycling condition was used for the primary amplification, with an initial denaturation step for 5 minutes at 95°C, five PCR cycles each at two different annealing temperatures (63°C and 56°C), and then 30 PCR cycles with an annealing temperature of 50°C. Each PCR cycle included a 40 seconds denaturation step at 95°C, a 40 seconds annealing step at the temperatures indicated above, and a 90 seconds extension step at 72°C and a final extension at 72°C for 10 minutes.
Mixture of nested secondary PCR included 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon), 1.0 μL of the 1:5 diluted PCR product described above, 1 μL of a gene-specific primer (5ACO1R3; Table 1), 1 μL of a primer complementary to the UPM long sequence (UPM short; Table 1) and 9.5 μL of ddH2O in a total volume of 25 μL. Touchdown cycling condition was used for the secondary amplification, with an initial denaturation step for 5 minutes at 95°C, five PCR cycles each at two different annealing temperatures (63°C and 56°C), and then 30 PCR cycles with an annealing temperature of 53°C. Each PCR cycle included a 40 seconds denaturation step at 95°C, a 40 seconds annealing step at the temperatures indicated above, and a 90 seconds extension step at 72°C and a final extension at 72°C for 10 minutes.
3.3. Multiple Sequence Alignment and Degenerate PCR
The 3ACO1F primer was designed from the fragment obtained by 5’ RACE method and degenerate PCR primer 3ACO1R was designed by the alignment and comparison of previously published ACO sequences from five Candida species; C. tropicalis, C. albicans, C. dubliniensis, C. parapsilosis, and C. orthopsilosis. All sequences were obtained from the GenBank (accession numbers XM_002548708, XM_711132, XM_002422275, HE605206, and XM_003869506 respectively). The PCR reaction mixture included 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon), 1 μL (10 pmole/μL) of each forward (3ACO1F) and reverse (3ACO1R) primers, 1 μL C. krusei genomic DNA and 9.5 μL ddH2O in a total volume of 25 μL. The cycling parameters were 95°C for 5 minutes: 30 cycles of denaturation for 40 seconds at 95°C, annealing for 40 seconds at 60°C, an extension for 90 seconds at 72°C, with a final extension of 10 minutes.
3.4. Cloning and Sequencing of PCR Products
All PCR products were cloned using a TOPO TA-cloning kit (Invitrogen, USA) following the manufacturer’s instructions. Products were ligated into the TA vector and then transformed into chemically competent cells using heat shock at 42°C. Cells were then mixed with 250 μL of SOC medium, incubated at 37°C for one hour, and then 50 μL of the cell culture was plated onto X-gal/IPTG/Amp/LB plates. Following overnight incubation at 37°C, a single white colony (~2 mm diameter) was picked from the plate and inoculated into 5 mL of Luria Bertani (LB) broth. After overnight incubation at 37°C, 1 mL of the culture was used to prepare glycerol stocks, and the other 4 mL was used to extract plasmid. Recombinant plasmid was purified using a plasmid extraction Kit (Intron biotechnology) following the manufacturer’s instruction. The recombinant plasmids were screened by M13 forward and reverse primers with the following PCR protocol: 94°C for 5 minutes; 30 cycles of 94°C for 1 minutes, 55°C for 1 minute; 72°C for 1 minute; and 72°C for 7 minutes. PCR products were analyzed by agarose (Merck, Germany) gel electrophoresis. The plasmids containing unknown DNA were sequenced by M13 primers (Bioneer, Korea).
3.5. Nucleotide and Protein Sequence Accession Number
The partial nucleotide sequence of the C. krusei aconitase gene and its annotation was submitted to the GenBank under the assigned accession number KJ009334.
3.6. Molecular Phylogenetic Analyzes
The following aconitase amino acid sequences from different organisms were obtained from the GenBank database. Candida tropicalis, XP_002548754.1; C. albicans, XP_716225.1; C. parapsilosis, CCE42927.1; C. orthopsilosis, XP_003869555.1; C. dubliniensis, XP_002422320.1; C. glabrata, XP_445684.1; Pichia pastoris, XP_002489444.1; Scheffersomyces stipitis, XP_001386080.1; Lodderomyces elongisporus, XP_001525166.1; Clavispora lusitaniae, XP_002619843.1; Piromyces sp. CAA76360.1; Schizosaccharomyces pombe, NP_594031.2; Penicillium marneffei, XP_002147388.1; Aspergillus oryzae, XP_001819597.2; A. flavus, XP_002374915.1; A. terreus, AAC61778.1; A. fumigatus, XP_750430.1; A. clavatus, XP_001269477.1; A. niger, XP_001393703.1; Trichophyton rubrum, XP_003231764.1; Arthrodermaotae, XP_002847793.1; A. gypseum, XP_003177439.1; Coccidioides posadasii, ABH10644.1; Paracoccidioides brasiliensis, XP_002790154.1; Ajellomyces dermatitidis, XP_002627881.1; A. capsulatus, XP_001539799.1; Cryptococcus gattii, XP_003193698.1; C. neoformans, XP_570245.1; Homo sapiens, CU012932.1.
Nucleotide and amino acid sequences were aligned using ClustalW online software. Sequences were analyzed by the Unweighted-Pair-Group method with arithmetic mean (UPGMA) and neighbor joining (NJ) using Geneious R7 software. Bootstrap probability with 2000 replications was introduced to assess the statistical significance of the groups in the phylogenetic tree.
3.7. Active Site and Binding Site Characterization
3D structure, active site and binding site residues of C. krusei ACO was identified with COFACTOR online service (http://zhanglab.ccmb.med.umich.edu/COFACTOR/) to determine active site and binding site amino acid residues differences between C. krusei ACO and other Candida species and human aconitases to design new antifungal drugs.
4. Results
In the current study, 1419 nucleotides of C. kruseiACO gene clarified the first 300 nucleotides of 3’ region of C. krusei ACO gene obtained by 5’-RACE method. 1197 nucleotides of this gene were identified by PCR with 3ACO1F primer as both forward and reverse primers, whereas the degenerate PCR primer designed by alignment and comparison of previously published ACO sequences from five Candida species was unable to amplify this gene (Figure 1). Seventy-eight primary nucleotides overlapped with the sequence obtained from the 5’-RACE method. Finally, sequenced nucleotides were submitted to the GenBank as a partial sequence of C. krusei ACO gene.
Table 2 exhibits the pairwise nucleotide identities of Candida species calculated from the nucleotide sequences of the ACO genes. Candida albicans and C. dubliniensis (96%), C. albicans and C. tropicalis (90%), C. dubliniensis and C. tropicalis (90%) and C. orthopsilosis and C. parapsilosis (94%) had more than 90% identity. Candida krusei had maximum identity with C. tropicalis (84%).It also shows the pairwise amino acid identities among different Candida species. Based on the amino acid sequences, C. krusei had maximum identity with C. tropicalis (85%); however C. albicans and C. dubliniensis (99%), C. albicans and C. tropicalis (94%), C. dubliniensis and C. tropicalis (94%) and C. orthopsilosis and C. parapsilosis (98%) had more than 90% identity.
| Species | Aconitase Sequence Similarity | |||||
|---|---|---|---|---|---|---|
| C. albicans | C. dubliniensis | C. tropicalis | C. parapsilosis | C. orthopsilosis | C. krusei | |
| Amino acid sequence similarity | ||||||
| C. albicans | ||||||
| C. dubliniensis | 96 | |||||
| C. tropicalis | 90 | 90 | ||||
| C. parapsilosis | 86 | 89 | 86 | |||
| C. orthopsilosis | 85 | 89 | 86 | 94 | ||
| C. krusei | 83 | 82 | 84 | 82 | 82 | |
| Nucleotide sequence similarity | ||||||
| C. albicans | 99 | 94 | 89 | 89 | 84 | |
| C. dubliniensis | 94 | 85 | 85 | 83 | ||
| C. tropicalis | 88 | 88 | 85 | |||
| C. parapsilosis | 98 | 81 | ||||
| C. orthopsilosis | 82 | |||||
| C. krusei | ||||||
aValues are reported based on percentage.
The comparison of Candida species ACO genes showed that intra species variations between C. krusei and other Candida species were relatively higher than the variation between common Candida species. Multiple alignments of the nucleotide (Figure 2) and amino acid (Figure 3) sequences of ACO of C. krusei with other Candida species and Homo sapiens as an out-group showed that C. krusei possessed characteristic sequences.
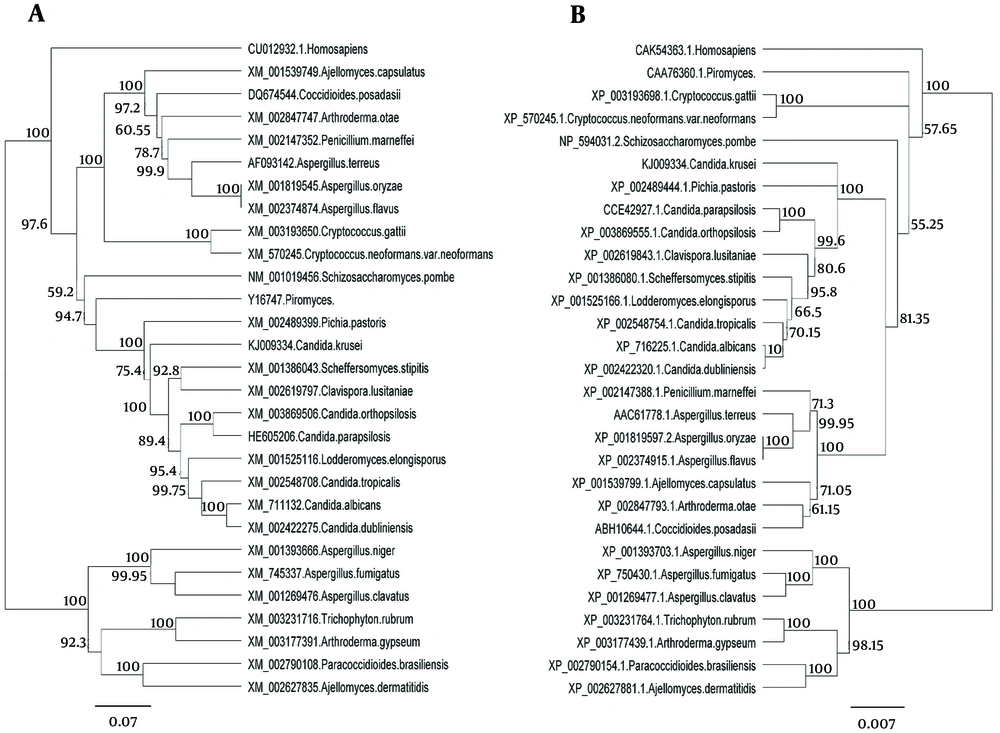
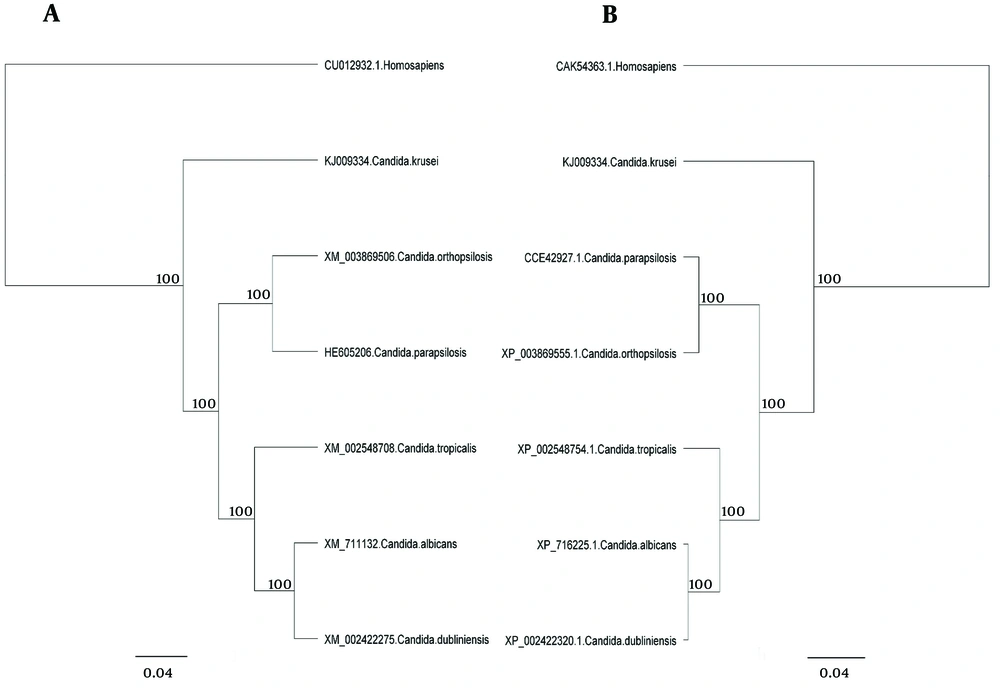
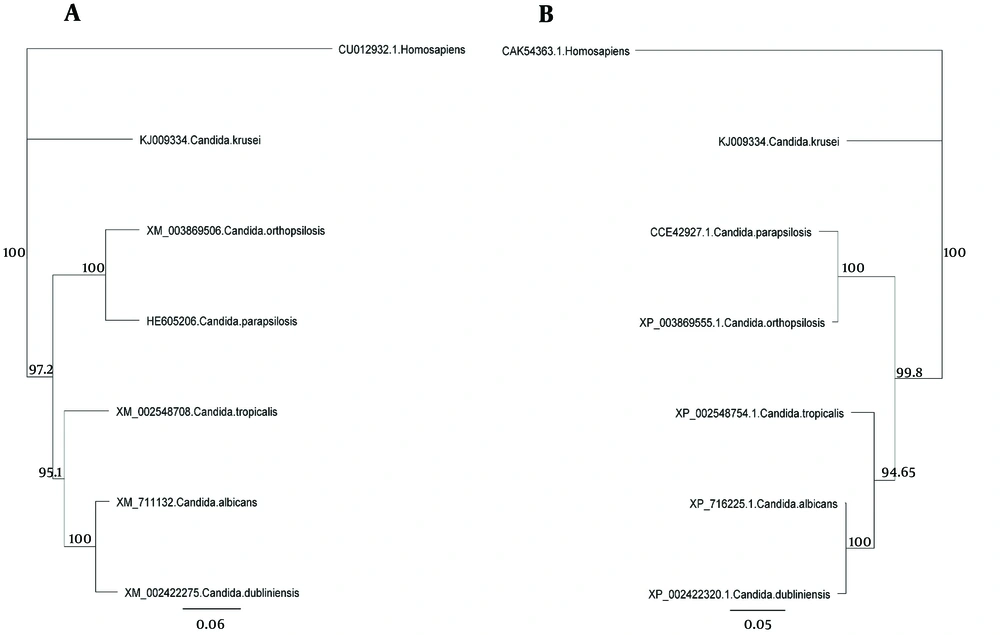
The major pathogenic Candida species were distinctly positioned by phylogenetic trees constructed using UPGMA and NJ with H. sapiens as an out-group (Figures 4 - 6). Although there were some difference in the degree of resolution when nucleotide and amino acid sequences were applied, the topologies acquired by both sequence types and tree estimation algorithms were consistent.
The active site and binding site of C. krusei aconitase contain six and ten residues, respectively (Table 3). The active site amino acid residues of ACO in six Candida species and human were similar; just the amino acids were located in different positions in protein structure. The binding site residues of ACO in six Candida species are identical but one isoleucine residue of C. kruseiACO was not present in aconitase binding site of human and there was one aspartate residue in human aconitase that was not present in C. krusei and other Candida species.
| Species | Active Site Residues | Binding Site Residues |
|---|---|---|
| C. krusei | Asp123, His124, His170, Asp188, His190, Glu285 | His124, Ile168, Ile169, His170, His190, Ser381, Cys382, Cys445, Cys448, Ile449 |
| C. albicans | Asp113, His114, His160, Asp178, His180, Glu275 | His114, Ile158, Ile159, His160, His180, Ser371, Cys372, Cys435, Cys438, Ile439, Asn460 |
| C. dubliniensis | Asp111, His112, His158, Asp176, His178, Glu273, Ser654 | His112, Ile156, Ile157, His158, His178, Ser369, Cys370, Cys433, Cys436, Ile437, Asn458 |
| C. tropicalis | Asp113, His114, His160, Asp178, His180, Glu275 | His114, Ile158, Ile159, His160, His180, Ser371, Cys372, Cys435, Cys438, Ile439, Asn460 |
| C. parapsilosis | Asp112, His113, His159, Asp177, His179, Glu274, Ser655 | His113, Ile157, Ile158, His159, His179, Ser370, Cys371, Cys434, Cys437, Ile438, Asn459 |
| C. orthopsilosis | Asp113, His114, His160, Asp178, His180, Glu275 | His114, Ile158, Ile159, His160, His180, Ser371, Cys372, Cys435, Cys438, Ile439, Asn460 |
| Homo sapiens | Asp114, His115, His161, Asp179, His181, Glu276, Ser656 | His115, Ile159, His161, Asp179, His181, Ser371, Cys372, Cys435, Cys438, Ile439, Asn460 |
5. Discussion
The current study aimed to identify C. krusei aconitase gene sequence and evaluate the phylogenetic relationships of C. krusei with other organisms and Candida species based on nucleotide and amino acid sequences of this gene. Also, the differences of active and binding sites and amino acid residues between C. krusei and human aconitase protein were determined to design new antifungal drugs targeting C. krusei aconitase. Since the length of this gene in Candida species is approximately 2340 base pair, it is believed that the length of this gene in C. krusei is also 2340 bp. In the current study, 1419 bp of aconitase gene of C. krusei were identified and sequenced by different methods including 5’ RACE method and degenerate PCR. Partial sequence was submitted to the GenBank and the accession number KJ009334 was assigned.
The 5’ RACE method in the current study was a new method introduced by Xian Zong Shi and Jarvis to identify GC rich sequences in 2006 (25). This method was slightly modified and used to identify a part of C. kruseiACO gene. Although the ACO gene of C. krusei is not GC rich but the results of this method to identify 5’ end of mRNA of this gene was satisfactory. Since 3’ RACE method was unable to identify the other part of this gene, degenerate PCR was used to identify unknown parts of ACO gene. Sequence analysis indicated that 1197 bp of this gene were amplified using PCR with 3ACO1F as both forward and reverse primers and degenerate PCR was unable to amplify this gene.
The ACO gene encodes protein aconitase involved in energy production in the Krebs cycle (19). Since this gene encodes a critical enzyme in different organisms including Candida species, little difference is expected between the nucleotide and amino acid sequences among different species of a genus. The results showed that not only nucleotide and amino acid sequences of this gene in C. krusei were very different from other species of Candida but also there were considerable differences between the other species of Candida. However, differences of nucleotide and amino acid sequences of this gene in C. krusei were higher than those of the other Candida species, which supports the hypothesis that C. krusei may be taxonomically separated from Candida genus. Phylogenetic analysis of amino acid and nucleotide sequences of aconitase gene showed that although C. krusei takes place in the Hemiascomycetes clade (26), this species takes place beside P. pastoris in phylogenetic trees and separated from other Candida species in this clade because of more nucleotide differences with other species (Figure 4).
The sequences were analyzed using UPGMA and NJ. There was very little difference between UPGMA (Figure 5), NJ (Figure 6), and phylogenetic trees based on the nucleotide or amino acid sequences of the common Candida species ACO genes. It seems to be due to bootstrap values and differences in the analysis methods. Moreover, different nucleotide codons of an amino acid may cause differences in nucleotide- and amino acid-based phylogenetic trees. Despite some differences in the degree of resolution when nucleotide and amino acid sequences were applied, the topologies obtained with both sequence types and the tree estimation algorithms were mutually consistent.
Aconitase enzyme activates in the presence of a [4Fe-4S] cluster (27). There is a [3Fe-4S] cluster in inactive aconitase ligated to the active site via three cysteine residues, Cys382, Cys445, and Cys448 in C. krusei (Cys372, Cys435, and Cys438 in human) and this enzyme is activated upon addition of a fourth iron atom (termed Fea) (28). This fourth iron atom has a slightly different electronic character than the others, and is the active site of the enzymatic reaction (28). There is no adjacent cysteine residue to ligate the fourth Fe (Fea), which instead binds a hydroxyl ion as its fourth ligand. The hydroxyl ion is bound to histidine (His) and aspartate (Asp) and becomes water (H2O) upon addition of substrate where it is further hydrogen bonded to the carboxyl group of citrate or isocitrate. Isocitrate or citrate is bound to Fea via an oxygen on its carboxyl group as well as its hydroxyl group, changing the coordination state of Fea from 4- to 6-coordinate (29). Coordination of the hydroxyl group makes it a better leaving group and facilitates the dehydration reaction that forms cis-aconitate from either citrate or isocitrate (27, 30). Serine (Ser) 656 alkoxide (in human) attacks the hydrogen on hydroxyl group of citrate or isocitrate creating a carbanion intermediate. Depending on which carboxyl oxygen coordinates the iron atom, cis-aconitate can be converted to citrate or isocitrate upon addition of water (30). In the TCA cycle, cis-aconitate is converted to isocitrate.
This is report the first report on the identification, sequencing and phylogenetic analysis of C. krusei using ACO gene sequences. Differences of C. kruseiACO gene sequences from other Candida species are prominent and may facilitate the design of probes for rapid and specific identification of C. krusei from other common Candida species. Moreover, according to aconitase activity mechanism, there are not significant amino acid residue differences between C. krusei and human aconitases within the active site cleft, which may block the use of aconitase as a molecular target applied to design new anti-fungal drugs that selectively block this enzyme.