1. Background
Neisseria meningitidis serogroup B (NmB) is an encapsulated Gram-negative organism and the cause of life-threatening bacterial meningitis worldwide, especially among infants and young adults (1-3). Unlike the other serogroups of N. meningitidis (A, C, Y and W135), the B capsular polysaccharide serogroup is similar to sialic acid of the foetal neural cell adhesion molecule (NCAM) and is poorly immunogenic; thus, there is no effective vaccine based on capsular polysaccharides against NmB (4-6). Therefore, to advance effective vaccines against NmB, outer membrane vesicle (OMV) proteins have been considered. Outer membrane vesicle-based vaccines have been used locally based on strain-specific OMV of NmB but the usage of these vaccines has been decreased by a variety of NmB strains, short period of immunogenicity, and lack of immune responses in young children (2, 5, 6).
Among nonpathogenic Neisseria species, N. lactamica shows close antigenic similarities with N. meningitidis. N. lactamica is a harmless commensal species and is more frequently colonized in the upper respiratory tract of infants and young children when compared to adults; its colonization rates decrease with increasing age (7-9). Previous studies have shown that development of cross-protective immunity against N. meningitidis will be seen in people who are carriers of N. lactamica. Therefore, it has been proposed that colonization with N. lactamica during early childhood will protect the host against colonization with N. meningitidis by natural immunity, which is thought to be due to shared common antigens (5, 7, 8, 10-12). Proteomics, as a key technology, is being widely used in the study of proteins using two-dimensional gel electrophoresis (2-DE), followed by mass spectrometric (MS) protein identification (13). With an aim to carry out N. meningitidis proteomics, Mignogna et al. published the proteome map of the NmB serogroup B strain MC58 (14).
Investigation of the proteomic profiles of the N. lactamica strains is rarely performed. Some proteomic investigations have been devoted to analyzing and comparing common outer-membrane vesicle (OMVs) proteins (12, 15) and OMVs vaccine antigens (16) between NmB and N. lactamica. Previous studies have reported that cytoplasmic proteins, in addition to outer membrane proteins, may be involved in immune responses, (17, 18). However, the whole proteome profiles of N. lactamica isolates are not well understood. Also, no comparison has been done on the cytoplasmatic and periplasmic proteome of N. lactamica and NmB, so far. Therefore, this proteomic approach is the first whole proteome analysis of N. lactamica strains. This study is also the first comparative whole proteome analysis of the N. lactamica and NmB strains for the comparison of their common cytoplasmic and membrane proteins using proteomics approaches.
2. Objectives
The main aims of this study were to determine whole proteome profiles of N. lactamica strains and to compare them with whole proteome profile of a reference strain of NmB, for identification of common cytoplasmic and membranous proteins between the proteins of the two species by two-dimensional electrophoresis (isoelectric focusing (IEF)/SDS-PAGE) (2DE) and subsequent matrix-assisted laser desorption/ionization-tandem time-of-flight mass spectrometry (MALDI-TOF/TOF-MS).
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
Four N. lactamica strains were used in this study. Three N. lactamica, Nl1, Nl2 and Nl3 were isolated from the oropharynx of healthy young children in Ahvaz, southwest of Iran (ethics number: ETH609). In our study, N. lactamica isolates, as gram-negative and oxidase-positive diplococci, were identified by growth on modified thayer-martin (MTM agar) medium (Oxoid, Basingstoke, UK) (19), Ο-Nitrophenyl-Β-D-Galactopyranoside disc (ONPG) test and polymerase chain reaction (PCR) for the presence of the pdhC gene (9). Identification of N. lactamica strains may be confirmed by detecting beta-galactosidase in an enzyme substrate test (16). Therefore, Β-galactosidase activity was tested with ONPG (Rosco Diagnostica, Denmark), as the substrate. Ο-Nitrophenyl-Β-D-Galactopyranoside disc test was performed according to the instructions of the manufacturer. The ONPG-positive isolates were yellow after incubation at 37°C for up to one hour (20). Neisseria lactamica ATCC 23970 and NmB ATCC 13090 were used as reference strains and obtained from the American type culture center, London, United Kingdom. All the bacteria were grown to confluence on Gonococcal (GC) agar (Oxoid, Basingstoke, UK) plus supplements at 37°C in a humidified atmosphere containing 5% CO2 (14, 21).
3.2. Whole-Cell Extract Preparation
Whole-cell protein extraction method was performed according to Bernardini (21). Briefly, the bacteria were harvested from three plates, resuspended in 2 mL of PBS plus an EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany), centrifuged at 8,000 × g (Eppendorf, Germany), for ten minutes at 4°C, and washed twice with the same buffer. The cell pellets were resuspended in 1.0 mL of reswelling lysis buffer: 7 M urea, 2 M thiourea, 2% (w/v) 3-((3-Cholamidopropyl)-dimethylammonio)- 1-proane sulphonate CHAPS, 2% (w/v) amidosulfobetaine-14 (ASB-14) (Sigma-Aldrich, USA), 2% (v/v) 3 - 10 Non-Linear (NL) Immobilized pH Gradient (IPG) buffer, 2 mM Tributylphosphine solution (TBP) (Sigma-Aldrich, USA) and 65 mM Dithiothreitol (DTT). Lysates were added with 1000 U of benzonase solution (Sigma-Aldrich, USA) and 2 mM MgSO4 (Merck, Germany) and incubated at room temperature for 30 minutes while shaking. The cell extracts were then centrifuged at 12,000 × g (Eppendorf, Germany) for two hours at 15°C and the supernatant was collected. The soluble protein extracts were stored at -80°C in single use aliquots. The total protein concentration was determined by the Bradford assay (22). Urea, thiourea, CHAPS, Immobilised pH Gradient (IPG) buffer pH 3 - 10 non-linear (NL), and dithiothreitol (DTT) were obtained from GE Healthcare Biosciences, USA.
3.3. Two-Dimensional Gel Electrophoresis (2-DE)
First dimension Isoelectric Focusing (IEF) was performed using the PROTEAN IEF Cell (Bio-Rad, USA). Approximately, 500 µg of protein extracts from each sample was brought to a final volume of 440 µl with reswelling buffer and a small amount of bromophenol blue (Merck, Germany) (21, 23). Next, proteins were adsorbed by active rehydration onto an Immobiline DryStrip (18 cm, pH 3 - 10 NL gradient; GE Healthcare, USA) by applying 50 V at 20°C for 16 - 18 hours. All IEFs were carried out at 20°C and focused at 10,000 V for 20 hours. Linear voltage was increased from 250 V to 5000 V during the first two hours, from 5000 V to 10,000 V for 4.5 hours until 70,000 V/hour was obtained at a maximum of 10,000 V. Before the second dimension, strips were equilibrated for 15 minutes in equilibration buffer (6 M urea, 30% v/v glycerol) (Merck, Germany), 2% w/v Sodium Dodecyl Sulfate (SDS) (GE Healthcare, USA), 0.05 M Tris-HCl (GE Healthcare, USA) pH 6.8) containing 65 mM DTT. This step was followed by a 15-minute incubation in equilibration buffer plus 2.5% (w/v) iodoacetamide (GE Healthcare, USA). The second dimension was run on a 12% acrylamide gradient gel (18 × 20 cm × 1.5 mm) with a constant current of 20 mA, using a Mini-PROTEAN 3 Cell (Bio-Rad, USA), until the dye reached the bottom of the gel. The gels were stained with colloidal Coomassie brilliant blue G-250 (Bio-Rad, USA), as previously described (24). The 2-DE was carried out in triplicates for each bacterial strain.
3.4. Image Analysis
The gels were scanned using a computing densitometer (GS-800 Imaging Densitometer, Bio-Rad, USA) at 400 dpi resolution and analyzed using the ImageMaster 2D Platinum software, version 7 (Melanie 7) (Swiss Institute of Bioinformatics, Switzerland). Spot quantification was determined according to the percentage volume (relative volume). Approximately, 800 reproducible protein spots were detected by comparing three replicas of the gels for each Neisserial strain. The reference image was set as a gel with the highest number of detected spots. Using match analysis, we matched the common spots automatically. The detected spots and matches were edited manually if necessary.
3.5. Matrix-Assisted Laser Desorption/Ionization-Tandem Time-of-Flight Mass Spectrometry Analysis
Protein spots, stained with colloidal Coomassie brilliant blue G-250, were cut from the gel using a pipette tip in a free keratin environment. The punched spots were transferred to microcentrifuge tubes (Greiner, Germany). Then, the spots were sent to the bioscience technology facility, department of biology, University of York, Wentworth Way, York, YO10 5DD, UK, for trypsin in-gel digestion and MALDI-TOF/TOF-MS analysis. Furthermore, MALDI-TOF/TOF-MS was performed using a Bruker ultraflex III MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany).
3.6. Database Search
The monoisotopic peptide masses created by MALDI-TOF/TOF MS were searched against the NCBInr protein database in the matrix science data base (MSDB) 20131226 and 20140122 using the MASCOT program (www.matrixscience.com) in the taxonomy group of Bacteria. A maximum of one trypsin missed cleavage per peptide, a peptide mass tolerance of ± 100 ppm and mass tolerance of ± 0.5 Da were allowed on the fragment. Other search parameters were carbamidomethylation of cysteine residues and oxidation of methionine residues, which were set as fixed and variable modifications, respectively. Peak intensity for each spot showed more than 60 peaks for peptide mass fingerprinting data. The matched peptides mainly demonstrated the strongest peaks. A maximum of 15 precursors were selected for fragmentation from each MS spectrum. In this study, MASCOT scores greater than 55 for all identified protein were indicated to be significant (P < 0.05). The theoretical Mr (molecular mass) and pI values (pH isoelectric) for each identified protein were calculated individually to ensure the quality of data, using the ExPASY-Compute pI-Mr tool (http://web.expasy.org/compute_pi/).
4. Results
4.1. Two-Dimensional Gel Electrophoresis Analysis
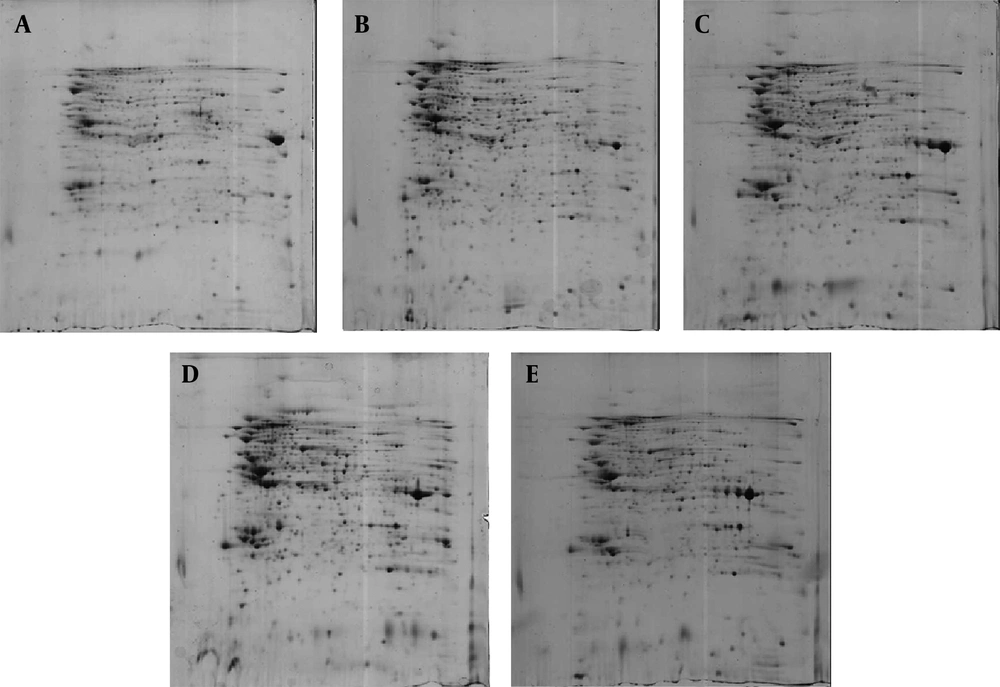
Bacterial strains were extracted in reswelling lysis buffer. Five hundred micrograms of any bacterial whole-cell lysates was separated by the use of 2-DE with Immobilized pH Gradient (IPG) strips (nonlinear pH 3 - 10) in the first dimension (isoelectric focusing). Using the ImageMaster 2D Platinum software, version 7 (Melanie 7), approximately 800 reproducible distinct protein spots stained with colloidal Coomassie brilliant blue G-250 from the two dimensional gels were visualized for each bacterial strain. Reproducibility was evaluated by comparing three replicas of the gels for each bacterial strain. For each gel, approximately 75% of the gel spots were matched according to the reference gel. Two-dimensional gel electrophoresis map of the most representative gel of the whole-cell extract of NmB and N. lactamica strains is shown in Figure 1. As shown in this Figure, by comparing 2-DE gels between NmB and N. lactamica strains, we found some of the variations in the proteome profile of the studied bacterial strains. The difference of the proteome profile between the studied N. lactamica strains was lower than NmB and N. lactamica strains. Therefore, after image analysis, we selected some of the common protein spots between the studied Neisseria strains from the 2-DE gel related to the reference strain of N. lactamica and some of the protein spots from the 2-DE gel related to the NmB for MOLDI-TOF/TOF mass spectrometry analysis.
4.2. Identification of proteins by Matrix-Assisted Laser Desorption/Ionization-Tandem Time-of-Flight Mass Spectrometry
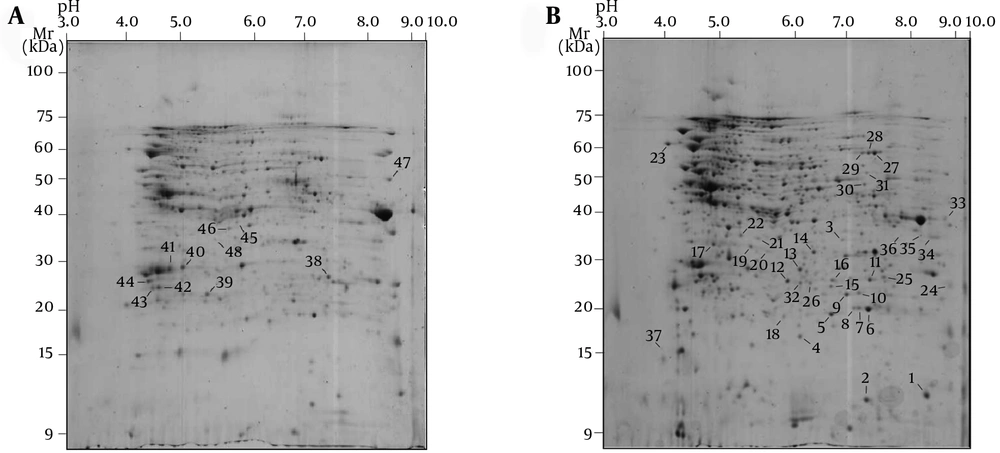
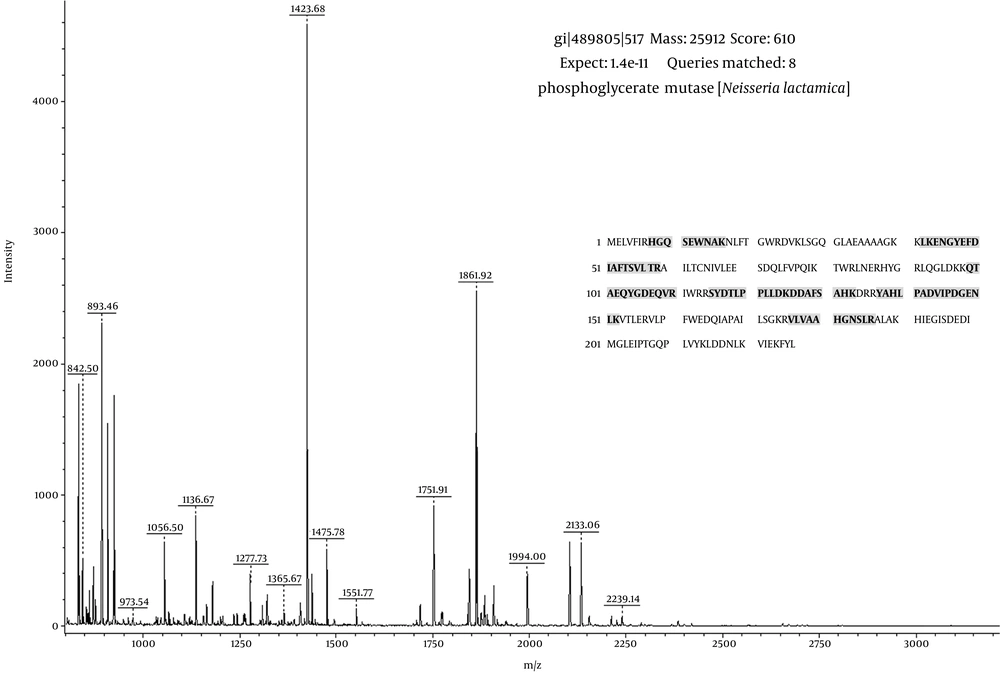
After image analysis, some of the protein spots were excised from the two 2-DE gels related to NmB and reference strain of N. lactamica and identified using MOLDI-TOF/TOF mass spectrometry. As shown in Figure 2, a total of 48 common protein spots were identified between NmB and N. lactamica strains, corresponding to 37 identified spot proteins in the N. lactamica strains and 11 identified spot proteins in NmB. Two of the protein spots (No. 7 and 38) did not significantly match using MALDI-TOF/TOF MS analysis. A list of MALDI-TOF/TOF MS-identified common protein spots between NmB and N. lactamica strains using the MASCOT program and the ImageMaster 2D Platinum software is reported in Table 1. As mentioned in Table 1, identified proteins ranged from theoretical pI 4.84 (spot 41) to 9.2 (spot 26) and from theoretical Mr 55 kDa (spot 23) to 15 kDa (spot 2). Other unidentified proteins were located between experimental pI 4.5-9.5 and experimental Mr of 9 - 100 kDa based on 2-DE gel image analysis. As shown in Table 1, in most identified proteins, a good agreement between theoretical and experimental pI and Mr was observed. The result of the peptide mass fingerprinting for spot number 15 is shown in Figure 3. As shown in Figure 3, the database search result using MASCOT and also protein sequence indicate the identity of the protein spot as phosphoglyceromutase. The standard used to accept protein spots identifications included the expanse of sequence coverage, the number of peptides matched, and the protein score. For most of the identified proteins, scores greater than 55 were considered significant (P < 0.05).
(A) 2-DE gel related to the NmB (B) 2-DE gel related to the reference strain of N. lactamica. Proteins corresponding to spot numbers are given in the Table 1.
| Spot | Protein Name | Cella | Mr (theor/exp) | pI (theor/exp) | Score | Matchedpeptides | Accession No. | Sequencecoverage, % | Function |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 50S ribosomal protein L10 | C | 17/11 | 8.8/7.7 | 272 | 2 | gi-489807274 | 27 | Translation |
| 2 | 50S ribosomal protein L9 | C | 15/10 | 6.6/7 | 384 | 3 | gi-313668457 | 36 | Translation |
| 3 | Pilus assembly protein PilW | U | 34/34 | 8.5/6.7 | 70 | 1 | gi-313668647 | 4 | Cell envelope (surface structures) |
| 4 | Single-stranded DNA-binding protein | C | 19/17 | 5.7/6.1 | 400 | 5 | gi-489806670 | 37 | Synthesis and modification of DNA |
| 5 | Superoxide dismutase | PS | 21/20 | 6.1/6.5 | 155 | 4 | gi-489803560 | 12 | Cell envelope (periplasmic proteins) |
| 6 | 50S ribosomal protein L25 | C | 21/20 | 7/7.4 | 431 | 4 | gi-489851556 | 31 | Translation |
| 7 | Not identified | /20 | /7.1 | ||||||
| 8 | 50S ribosomal protein L25 | C | 21/20 | 7/7 | 187 | 2 | gi-489851556 | 13 | Translation |
| 9 | Acetyl-CoA carboxylase carboxyl transferase subunit | C | 35/23 | 6.4/7.2 | 575 | 6 | gi-313668288 | 29 | Fatty acid biosynthesis |
| 10 | 3-ketoacyl-ACP reductase | U (ps) | 25/21 | 6.6/7.1 | 591 | 6 | gi-313669143 | 34 | Fatty acid biosynthesis |
| 11 | Uridylate kinase | C | 25/21 | 6.7/7.4 | 458 | 5 | gi-488172473 | 30 | Purines, pyrimidines, nucleosides and nucleotides biosynthesis |
| 12 | Ferredoxin--NADP reductase | C | 29/21 | 5.7/5.4 | 239 | 3 | gi-15676930 | ||
| 12 | Energy metabolism (respiration) | ||||||||
| 13 | Enoyl-ACP reductase | CM | 27/21 | 5.6/5.7 | 312 | 3 | gi-518101971 | 14 | Fatty acid biosynthesis |
| 14 | CRISPR-associated protein Csh2 | C | 32/22 | 5.8/6.1 | 590 | 7 | gi-489804777 | 32 | Synthesis and modification of macromolecules (DNA and RNA) |
| 15 | Phosphoglyceromutase | C | 25/21 | 6.1/6.5 | 610 | 7 | gi-489805517 | 37 | Energy metabolism (glycolysis) |
| 16 | Short-chain dehydrogenase | C | 26/21 | 6/6.5 | 164 | 2 | gi-489803682 | 11 | Energy metabolism (respiration) |
| 17 | Thioredoxin reductase | U (ps) | 33/22 | 5.1/3.7 | 412 | 4 | gi-518101583 | 21 | Purines, pyrimidines, nucleosides and nucleotides metabolism |
| 18 | Oxidoreductase | C | 20/20 | 5.6/5.3 | 205 | 2 | gi-489806338 | 15 | Miscellaneous |
| 19 | Phosphoribosyl-aminoimidazole-succinocarboxamide synthase | C | 32/22 | 5.4/4.6 | 507 | 5 | gi-489804157 | 25 | Purines, pyrimidines, nucleosides and nucleotides biosynthesis |
| 20 | Hypothetical protein | C | 33/22 | 5.5/4.8 | 169 | 2 | gi-313668735 | 10 | hypothetical protein |
| 21 | Methylenetetrahydrofolate dehydrogenase/cyclohydrolase | U (ps) | 30/23 | 5.4/4.7 | 447 | 5 | gi-489804966 | 26 | Biosynthesis of cofactor, prosthetic groups and carriers (folic acid) |
| 22 | NAD synthetase | C | 30/23 | 5.6/4.3 | 301 | 4 | gi-489803893 | 19 | Synthesis and modification of macromolecules |
| 23 | Nutilization substance protein A | C | 55/27 | 4.5/2.6 | 531 | 6 | gi-313667982 | 15 | Synthesis and modification of macromolecules (RNA and DNA) |
| 24 | Opa protein | OM | 26/23 | 9.2/9.3 | 184 | 2 | gi-8346997 | 12 | Cell envelope (surface structures) |
| 25 | Competence lipoprotein | OM | 30/25 | 8.6/7.6 | 250 | 3 | gi-313668817 | 13 | Transport/binding protein |
| 26 | ABC transporter | CM | 27/25 | 5.7/6.3 | 79 | 1 | gi-518102710 | 5 | Transport/binding proteins (cations) |
| 27 | Inosine-5’-monophosphate dehydrogenase | C | 52/53 | 6.7/7.4 | 384 | 4 | gi-313668309 | 12 | Purines, pyrimidines, nucleosides and nucleotides biosynthesis |
| 28 | Inosine-5’-monophosphate dehydrogenase | C | 52/53 | 6.7/7.2 | 513 | 6 | gi-313668309 | 19 | Purines, pyrimidines, nucleosides and nucleotides biosynthesis |
| 29 | Inosine-5’-monophosphate dehydrogenase | C | 52/53 | 6.7/7 | 75 | 1 | gi-313668309 | 3 | Purines, pyrimidines, nucleosides and nucleotides biosynthesis |
| 30 | Peptidoglycan-binding protein LysM | U | 45/47 | 8.3/6.9 | 314 | 3 | gi-489810301 | 10 | Cell envelope |
| 31 | Transcription termination factor rho | U (c) | 38/49 | 6.8/6.9 | 98 | 1 | gi-3421248 | 6 | Synthesis and modification of macromolecules (RNA and DNA) |
| 32 | Glutamine ABC transporter ATP-binding protein | CM | 26/24 | 5.5/6.2 | 65 | 1 | gi-489804998 | 4 | Transport/binding protein |
| 33 | Polyphosphate kinase 2 | C | 36/37 | 8.6/8.7 | 370 | 4 | gi-313669185 | 18 | Energy metabolism |
| 34 | Imidazoleglycerol-phosphate dehydratase | C | 33/34 | 7.8/8 | 273 | 4 | gi-313668038 | 16 | Histidine metabolism |
| 35 | Acetyl-CoA carboxylase carboxyl transferase subunit | C | 35/33 | 6.4/7.5 | 403 | 3 | gi-313668288 | 16 | Fatty acid biosynthesis |
| 36 | Porin | OM | 31/34 | 6.7/7.1 | 115 | 1 | gi-1669708 | 4 | Cell envelope (surface structures) |
| 37 | Inorganic pyrophosphatase | C | 19/20 | 4.8/4.7 | 77 | 1 | gi-313668872 | 5 | Lipid metabolism ( synthesis and degradation), calcium absorption and bone formation, DNA synthesis, and other biochemical transformations |
| 38 | Not identified | /26 | /7.2 | ||||||
| 39 | DNA-binding response regulator | C | 24/24 | 5.4/5.6 | 202 | 2 | gi-15676500 | 16 | modification of macromolecules (DNA) |
| 40 | Macrophage infectivity potentiator | OM | 28/27 | 5.7/5.3 | 302 | 3 | gi-15677418 | 14 | Surface structures (virulence factor) |
| 41 | Macrophage infectivity potentiator | OM | 28/27 | 5.7/5.1 | 384 | 4 | gi-15677418 | 18 | Surface structures (virulence factor) |
| 42 | Amino acid permease substrate-binding protein | PS | 28/24 | 5.6/5 | 620 | 6 | gi-121634581 | 29 | Transport/binding protein (amino acids and amines) |
| 43 | Putative disulfide oxidoreductase | PS | 25/24 | 5.4/4.9 | 181 | 2 | gi-6900471 | 10 | Miscellaneous |
| 44 | Amino acid ABC transporter substrate-binding protein | PS | 29/25 | 5/4.8 | 634 | 5 | gi-15677462 | 37 | Transport/binding proteins (cations) |
| 45 | Ketol-acid reductoisomerase | C | 36/37 | 5.6/6 | 579 | 6 | gi-15677424 | 26 | Amino acid biosynthesis |
| 46 | D-lactate dehydrogenase | C | 36/35 | 5.7/5.8 | 262 | 4 | gi-15677533 | 14 | Energy metabolism (respiration) |
| 47 | Outer membrane protein P1 | OM | 50/46 | 9.3/8 | 255 | 3 | gi-15676020 | 9 | Cell envelope (surface structures) |
| 48 | Succinyl-coa synthetase subunit alpha | C | 30/33 | 5.7/5.6 | 170 | 2 | gi-488142532 | 10 | Energy metabolism (tricarboxylic acid cycle) |
Matrix-Assisted Laser Desorption/Ionization-Tandem Time-of-Flight Mass Spectrometry Identification of Common Protein Spots Between Neisseria meningitidis serogroup B and Neisseria lactamica Strains
According to the mass spectrometry (MS) analysis, the identified proteins included the cytoplasmic, periplasmic, cytoplasmic, and outer membrane proteins (Table 1). The predicted cellular localization was obtained using the PSORTb program v.0.3 (25) with a cut-off value of 7.5. When all of the localization scores were equally presumable, the notation U (for unknown) was selected; this indicates that the protein may have multiple localization sites.
5. Discussion
The identification of the N. lactamica whole proteome can be used for the determination of common proteins among N. lactamica, as a harmless commensal species, and NmB, as an obligate human pathogen, to develop an effective vaccine based on their common proteins, to prevent life-threatening bacterial meningitis. Investigation of the proteomic profiles of the N. lactamica strains and their comparison with proteome profiles of NmB strains should be considered, because there are only a few studies that have been conducted in this regard. There is no information about N. lactamica strains whole-cell proteome profile, already available for NmB strain MC58 (14). However, some studies have been performed, analyzing the outer membrane proteins (12, 15) and outer membrane vesicle vaccine antigens (16) in N. lactamica and NmB. For this reason, there is great interest in identification of N. lactamica proteome profile. In this study, the whole-cell extracts of NmB and N. lactamica strains were used for comparative proteomic analysis to identify common proteins between the two species by 2-DE and MOLDI-TOF/TOF-MS analysis.
We compared three replicas of the 2-DE gels for each of the studied bacterial strain. Most of the 800 protein spots yielded in any of the 2-DE gels were located in the range of pI 4.5 - 9.5 and Mr of 8 - 100 kDa. In our study, the proteins with Mr of more than 100 kDa were not separated in 2-DE gels because previous studies have shown that some of the proteins with high molecular masses, and highly hydrophobic proteins, particularly membrane proteins, may not be separated due to solubilization problems during the Isoelectric focusing (IEF) step and may not enter the gels (26-28). In contrast to the results of this study indicating Mr of 8 - 100 kDa, Abel et al. demonstrated Mr of 16 - 80 kDa in their study (12). These studies revealed that all membrane proteins still cannot be properly solubilized and separated using a unique method. In this study the theoretical molecular masses and pI values of the identified protein spots were very close to those values observed in the 2-DE gel. In our study, as shown in Table 1, among the identified protein spots, spot No. 47 (outer membrane protein P1) with theoretical Mr of 50 kDa was the largest membranous protein. In this study, the studied N. lactamica strains had approximately similar proteome to each other in the total number of spots and the general profile. The 2-DE gels showed that whole proteome maps of N. lactamica strains and NmB were slightly different. Consistent with this study, Abel et al. reported that different strains might show different proteome profiles despite having the same protein composition (12). In this study we did not aim to carry out identification of different protein spots between the studied bacterial strains.
In the current study, most of the protein identified spots were based on ≥ 2 matched peptides, although some of them were based on a single matched peptide (spots No. 3, 26, 29, 31, 32, 36 and 37). However, protein scores for all the identified protein spots were higher than the significance threshold (P < 0.05). According to the results obtained from peptide mass fingerprinting, among the identified protein spot sequences, 11 and 18 spots from the 2-DE gel, related to the reference strain of N. lactamica, matched with proteins belonging to N. lactamica ATCC 23970, and N. lactamica, respectively. Ten protein spot sequences from the 2-DE gel, related to the NmB, matched with proteins belonging to N. meningitidis MC58.
More than 200 common proteins were recognized between the two species in 2-DE gels. The analysis of 48 identified protein spots by using PSORTb program revealed that 19 protein spots were membrane proteins including periplasmic, cytoplasmic and outer membrane proteins. In contrast to our study, Abel et al. identified 17 common membrane protein spots in N. lactamica strains (12). Also, they detected a small number of common membrane proteins between NmB and N. lactamica strains. Among the identified outer membrane proteins, outer membrane protein P1, porin, Opa and competence lipoprotein were reported as common outer membrane proteins between the two species in previous studies, with the results being in agreement with the present study (15, 16). The PSORTb program showed that some proteins may have multiple localization sites in the cell, for example peptidoglycan-binding protein LysM (spot No. 30), as a membranous protein, may be present at any location within the cell membrane. Other identified protein spots were cytoplasmic proteins.
Eleven protein spots (No. 38 - 48) from the 2-DE gel of NmB were identified, as shown in Table 1. Comparison of the 2-DE gel of two NmB and the reference strain of N. lactamica revealed that these 11 identified proteins were common proteins between the two bacterial strains mentioned above. Similar to our results, amongst the above-mentioned 11 identified protein spots, four spots (No. 39, 45, 47, 48) were reported by Mignogna et al. (14). Spots No. 40, 41, 42, 43, 44, and 46 were identified only by the present study. Spot No. 38 had no significant match and was not identified. Investigation of relative volume (%) of identified protein spots revealed that most of the protein spots, but not all, had similar spot volume in 2-DE gels. Some of the protein spots (spots No. 12, 13, 14) in the 2-DE gels related to NmB have a lower spot volume than the same spots in 2-DE gels related to N. lactamica strains.
The results indicated that the protein spots identified by MOLDI-TOF/TOF mass spectrometry, included cell surface, energy metabolism, amino acid transport and metabolism, coenzyme metabolism, defense, multifunctional, cellular processes, DNA, RNA and protein synthesis, ribosomal structure, regulatory functions, replication, transcription, translation, unknown and hypothetical proteins with unknown function. Cellular localization of these proteins in the cytoplasm, periplasm, cytoplasmic and outer membrane is shown in Table 1. The availability of genome sequences combined with protein databases of NmB strain MC58 (14) has led to research on the meningococcal proteome for developing an effective vaccine against this important human pathogen. In this context, it is important to consider that for performance of a reliable proteomics approach for N. lactamica strains and also NmB ATCC 13090, we must have the genome sequences of these bacteria. Therefore, this study focused on comparative proteome profiles and identification of some of the common proteins between non-pathogenic strains of N. lactamica and a pathogenic strain of NmB.
In conclusion, we identified several common proteins between N. lactamica strains and NmB using 2-DE-based MS analysis. Some of our identified proteins, but not all, were in line with previous studies that used 2-DE and MS analysis. This proteomic approach provides us with earlier reports of whole proteome maps of N. lactamica strains. Finally, we suggest that further studies are required to compare proteomics of this concept especially when genome sequencing of N. lactamica is still in progress.