1. Background
The family Anelloviridae contains eleven genera of human and animal viruses that have a single-stranded, circular, negative sense DNA genome (1, 2). In 2007, Ninomyia et al. identified a novel DNA virus with a genomic organization resembling those of TTV and TTMV and designated this new virus as Torque Teno Midi Virus (TTMDV), they also showed that Small Anellovirus (SAV1) and SAV2 were deletion mutants or artifacts generated during the amplification of TTMDV (3). Therefore, these viruses were named TTMDV/SAV and then TTMDV viruses and were classified as Gammatorquevirus genus (including 15 species: Torque teno midi virus 1 - 15) of the family (1-4).
TTMDV/SAV DNA was found in 20% of plasma samples of French blood donors, which 33% of their saliva samples were positive too (5). TTMDV/SAV DNA was detected in 9.1% of serum samples of patients with hepatitis C infection and 8.6% of healthy individuals in Italy (6). In Korea, TTMDV/SAV was found in 34% of blood products and 40% of patients with chronic hepatitis (7). Also in another study, TTMDV/SAV was detected in the serum of healthy children, children with acute respiratory disease (ARD), hepatitis B and hepatitis C, and nasopharyngeal aspirates collected from children with ARD (8). TTMDV/SAV is reported, in 46.7% of nasopharyngeal aspirates collected from children with acute respiratory disease in Hungary (1) and in 50.4% of different cervical cancers of the female Iranian population (9). The role of TTMDV /SAV in clinical diseases and the full geographical distribution of the virus is unclear (1).
High levels of TTV DNA and TTMV DNA are reported in the bone marrow and spleen tissues of HIV positive individuals and HIV positive individuals who had progressed to acquired immune deficiency syndrome (AIDS) (10). TTV and TTMV loads increase in immune suppression and reduce the number of CD4+ T cells (10, 11). Viral hepatitis is common and considered as a serious public health problem (12, 13). Also, it is the cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma worldwide (14-16). As these viruses share the same transmission routes such as blood transfusion, co-infection of TTMDV/SAV and Hepatitis B Virus (HBV), or TTMDV/SAV; Hepatitis C Virus (HCV) is expected. However, the effect of TTMDV/SAV infection in patients with chronic HBV or HCV infection is unknown.
2. Objectives
Considering the possible role of other members of Anelloviridae as etiological factors in hepatitis cases (17, 18) and that TTMDV/SAV virus is transmitted through blood products and can infect humans (8), the current study aimed to detect the frequency of TTMDV/SAV in the sera of HIV/HCV, HIV infected, and healthy individuals and their possible correlation, and also the possible role of TTMDV/SAV in the liver diseases was investigated.
3. Materials and Methods
3.1. Sampling and Serological Tests
Fifty-two, thirty-six, four, and one hundred-ten serum samples from HIV positive, patients with HIV/HCV and HIV/HCV/HBV co-infected cases regardless of the presence of clinical signs or not, and healthy individuals (not positive for HIV, HBV and HCV, and also normal routine blood tests such as counting blood cells (CBC) and liver enzymes) were collected from pathobiology laboratories in Khorramabad from December 2010 to September 2011, respectively. Patients’ characteristics, gender, and age were recorded. All samples were stored at -20°C until tested. The study was approved by the local University of Isfahan ethics committee. Different enzyme linked immunosorbent assay (ELISA) tests were used to detect anti-HIV antibody (Delaware Biotech, USA), HBs antigen (Delaware Biotech, USA) and anti-HCV antibody (DIA. PRO, Italy). Serum aminotransferases (AST and ALT) were evaluated by Biology Chemistry kit (Iran).
3.2. Nucleic Acid Extraction From Serum
For each sample 220 μL of serum was mixed with 6.5 μL of 0.25% Sodium Dodecyl Sulfate (SDS) and 10 μL of 0.2 M NaCl. Then, 12 μL of 10 mg/mL proteinase K (Roche, Germany) was added and thermo-mixed at 350 rpm for 2 hours at 65°C. Proteins were precipitated using phenol and chloroform. The supernatant phase containing DNA was collected and transferred to a microtube and 3 M sodium acetate was added (0.1 volume). Then, the samples were vortexed for 30 seconds. DNA was precipitated with the cold ethanol (100%) (Merck, Germany). The pellet was dried and dissolved in 50 μL of distilled deionized water or TE (Tris-HCL buffer). To determine the accuracy of the extracted DNA, it was electrophoresed on 1% agarose gel containing ethidium bromide and studied under UV light. To determine the purity of the extracted DNA, absorbance of extracted DNA was read at wavelengths of 260 and 280 nm by the spectrophotometer.
3.3. Detection of TTMDV/SAV DNA by Nested-PCR
The SMAs1/SMAr1 and SMAs2/SMAr2 primer sets (5, 7) were used to perform nested Polymerase Chain Reaction according to Salmanizadeh et al. (9). The sequences of TTMDV/SAV virus specific primers were: SMAs1: GGCGGGAGCCCGAGGTGAGTGAAAC (sense), SMAr1: TTCTAATAGAGTCTGGTTGCCATTG (antisense), SMAs2: ACCGAGGTCTAGGGGCAATTCGGGC (sense), and SMAr2: CCGGCGGCGTCTTCTGCGGCGGCGA (antisense). A positive control was used, which was already sequenced with accession number of HQ259681 in the GenBank (9).
3.4. DNA Sequencing
PCR products of seven randomly selected samples from patients with HIV/HCV and HIV/HCV/HBV co-infections, and healthy blood donors were subjected to agarose gel electrophoresis (1.5%) and DNA was extracted according to the instructions of the DNA gel extraction kit K0513 (Fermentas, Germany). The DNAs were sequenced by Takapouzist Company, Iran. BLAST Nucleotide search of the determined sequences against the nucleotide sequence database (NCBI, national center for biotechnology information) was performed.
3.5. Molecular Evolutionary Analysis
Phylogenetic trees were constructed using neighbor-joining method in MEGA4 (Molecular Evolutionary Genetics Analysis software version 4.0) based on the current study sequenced amplicons against sequences obtained from the GenBank (19).
3.6. Statistical Analyses
The Fisher exact test and paired t-test were employed for statistical analyses using the GraphPad Instat software version 3.05 (GraphPad software, USA). P value < 0.05 was considered statistically significant.
4. Results
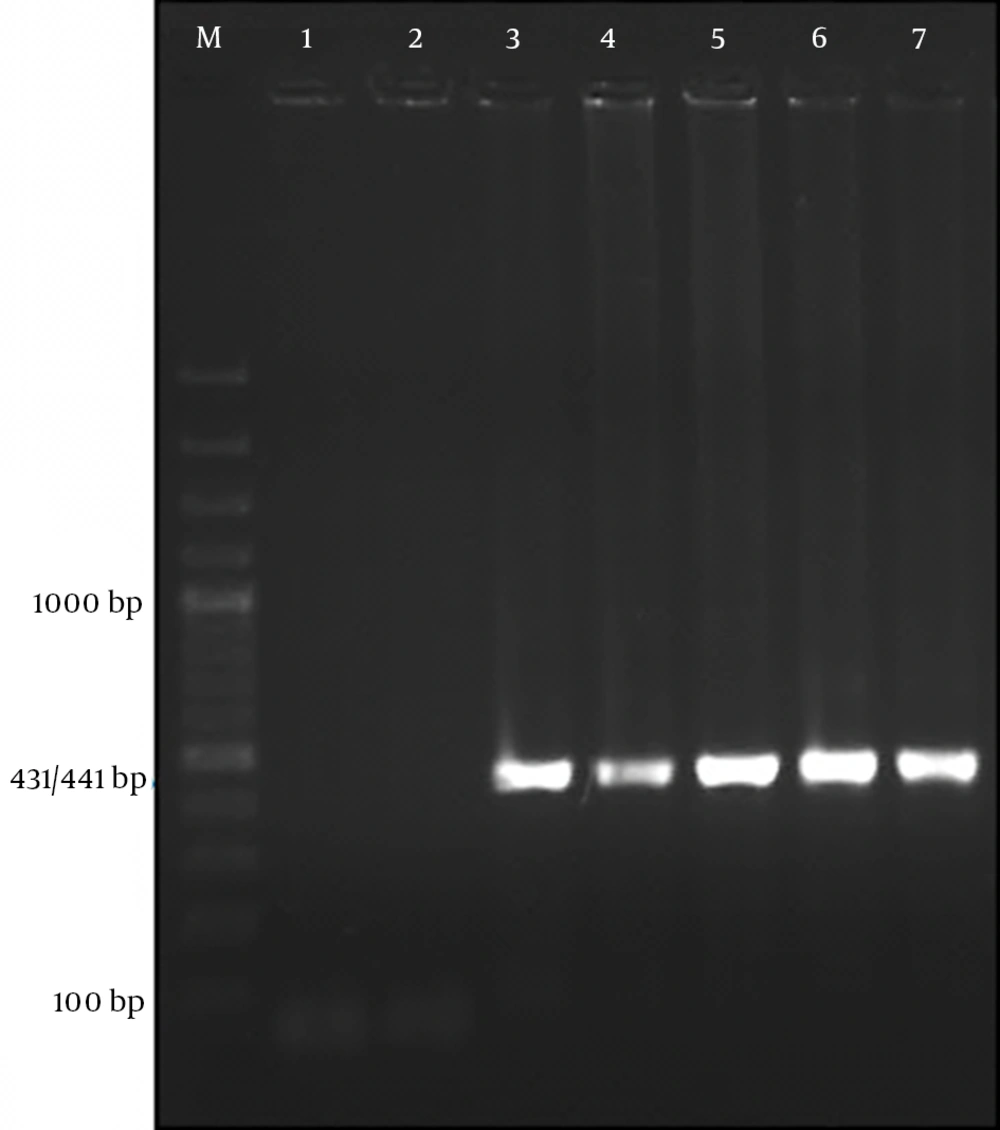
In the gel electrophoresis expected 431 base pair product for SAV2 and 441 bp product for SAV1 (4, 8) were observed (Figure 1). Alignment of the seven obtained sequences (GenBank accession numbers: KC352905, KC352906, KC352907, KC352908, KC352909, KC352910, KC352911) revealed a high degree of genetic variability among the TTMDV/SAV DNA (homology ranging from 70% to 93%) with some sequences in the Gen Bank. As indicated in Figure 2, three of the sequences related to patients with HIV/HCV co-infection (KC352906, KC352908, and KC352910) were located in a separate cluster and had less similarity with other reported cases. Although the other four sequences (KC352905, KC352907, KC352909, and KC352911) showed differences, they were generally more similar to the other reported sequences. Also, as shown in Figure 3, there was high degree of genomic homology of the sequences with some SAV1 sequences (sequences with accession numbers DQ987836 and DQ409192) and SAV2 sequences (sequences with accession numbers DQ987853, DQ987851, and DQ987854) reported by Chung et al. (20).
The evolutionary distances were computed using the maximum composite likelihood model based on the units of the number of base substitutions per site. KR-HB (Korean patients with hepatitis B virus infection), KR-CT (Korean control cases), KR-HC (Korean patients with hepatitis C virus infection), GenBank accession numbers of the sequences from top to bottom of the figure (DQ978799, JN112516, DQ978803, AY622909, DQ978792, DQ978795, NC-009225, AY622908, NC-014093, JN112517, KC352911, DQ978801, DQ978802, KC352905, KC352909, DQ978796, DQ978793, KC352907, HQ259681, DQ978798, DQ978791, DQ978794, HQ259682, KC352906, KC352908, KC352910 and DQ186999).
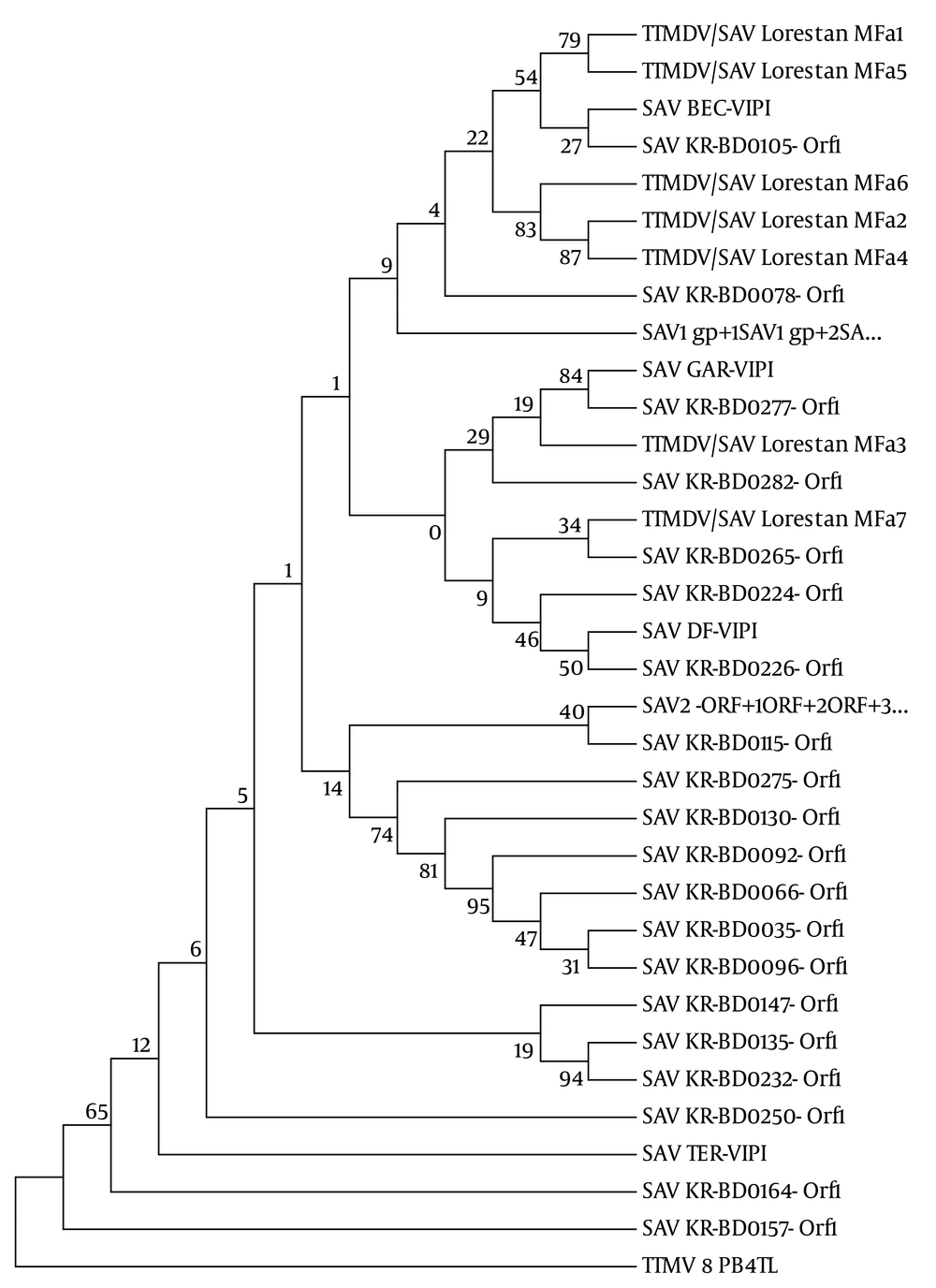
The evolutionary distances were computed using the maximum composite likelihood model based on the substitutions per site. GenBank accession number: Small anellovirus 1 (SAV1 gp1 + SAV1 gp2 + SA: NC-007013), Small anellovirus 2 (SAV2 ORF1 + ORF2 + ORF3 + ORF4 + …: AY622909), SAV DF-VIPI (S. anellovirus DF-VIPI: DQ409193), SAV BEC-VIPI (S. anellovirus BEC-VIPI: DQ409192), SAV GAR-VIPI (S. anellovirus GAR-VIPI: DQ409196), SAV TER-VIPI (S. anellovirus TER-VIPI: DQ409199), SAV KR-BD (Korean blood donors: DQ987831-DQ987854), Torque teno mini virus (T. teno mini virus 8 PB4TL: AB038631), and our sequences Torque teno midi virus/small anellovirus Lorestan MFa1 (patients with HIV/HCV/HBV co-infection: KC352905), Torque teno midi virus/small anellovirus Lorestan MFa3 (healthy individual: KC352907), Torque teno midi virus/small anellovirus Lorestan MFa2, MFa4, MFa5, MFa6, MFa7 (patients with HIV/HCV infection: KC352906, KC352908-KC352911).
Features and information of serum samples from blood donors are shown in Table 1. In the current study, healthy blood donors were used as the control group. The frequency of TTMDV/SAV in different serum samples are shown in Table 2. The differences of the TTMDV/SAV frequencies in the control cases versus HIV positive and HIV/HCV co-infected cases were statistically significant (P < 0.01). The difference between HIV positive and HIV/HCV co-infected cases was not significant (P > 0.05).
| Sample | Number | Gender (Male/Female) | Mean Age |
|---|---|---|---|
| Healthy blood donors | 110 | 52/58 | 25.8 |
| HIV positive | 52 | 26/26 | 38.25 |
| HIV/HCV co-infected | 36 | 35/1 | 38 |
| HIV/HCV/HBV co-infected | 4 | 4/0 | 42.75 |
Serum Samples Information
| Sample | TTMDV/SAV Positive | TTMDV/SAV Negative |
|---|---|---|
| Healthy | 16 (14.5) | 94 (85.5) |
| HIV positive | 43 (82.7) | 9 (17.3) |
| HIV/HCV co-infected | 29 (80.5) | 7 (19.5) |
| HIV/HCV/HBV co-infected | 3 (75) | 1 (25) |
| Total | 91 (45) | 111 (55) |
Frequency of Torque teno midi virus in the Serum Samplesa
There were not any significant differences in the means of the serum aminotransferases (ALT and AST) levels between TTMDV/SAV-positive and TTMDV/SAV-negative individuals in different groups (P > 0.05). Due to the low number of HBV infected samples, comparison was not possible (Table 3). Also in each age group the level of ALT and AST were not significantly different between the patients with and without TTMDV/SAV infection (P > 0.05). Comparisons of different age groups between the patients with and without TTMDV/SAV infection in healthy and HIV positive and HIV/HCV co-infected patients are shown in tables 4 and 5, respectively. About 46.5% of HIV positive patients and 55% of those with HIV/HCV infection, co-infected with TTMDV/SAV, were in the age group of 31 - 40 years. The highest prevalence of healthy blood donors was 56% which was in the age group of 20 - 30 years. Prevalence of TTMDV/SAV was not significantly different between males and females, in the healthy blood donors and the HIV positive groups (P > 0.05).
| Characteristicsb | TTMDV/SAV (+) | TTMDV/SAV (-) | P Value |
|---|---|---|---|
| AST of HIV positive individuals | 79.41 ± 8.92 | 53.88 ± 8.92 | 0.2051 |
| ALT of HIV positive individuals | 64.28 ± 7.46 | 47.22 ± 9.17 | 0.628 |
| AST of HIV/HCV co-infected individuals | 75.27 ± 8.53 | 110.7 ± 36 | 0.367 |
| ALT of HIV/HCV co-infected individuals | 68.9 ± 8.3 | 100.14 ± 31.9 | 0.479 |
| AST of HIV/HCV/HBV co-infected individuals | 99 ± 30.5 | 129 ± 0 | - |
| ALT of HIV/HCV/HBV co-infected individuals | 86.66 ± 26.66 | 110 ± 0 | - |
Comparison of Aspartate Aminotransferase and Alanine Aminotransferase Levels in the Serum Samples With and Without Torque teno midi virus/Small anelloviru virus Infectiona
The Distribution of Infection Among Different Age groups in Healthy Individuals and Patients With HCV/HIV Co-Infection
5. Discussion
The current study results revealed a high prevalence of the virus in the population of the Lorestan province. The high frequency of the virus in the cases with hepatitis was much higher than the cases reported elsewhere. In the present study, high frequency of TTMDV/SAV in HIV positive individuals and those with HIV/HCV co-infection shows the possibility of association between TTMDV/SAV and blood-borne infections such as HIV and hepatitis C, and also indicates the main route of blood transfusion of the virus rather than via saliva, infected peripheral blood mononuclear cells (5), the respiratory tract (8), nasopharyngeal discharge, urine, stool (1), and sexual transmission (9) already reported. According to Garcia-Alvarez et al. TTV and TTMV may cause liver diseases in patients co-infected with HIV and HCV (18). High frequencies of TTMDV/SAV among patients with HIV/HCV co-infection in comparison with low frequencies in healthy ones suggest that the virus is highly compatible with blood-borne viruses. This may lead to the greater stability of the virus in the patients with HIV/HCV co-infection.
This is the first time that TTMDV/SAVs are detected in HIV positive individuals in Iran and worldwide. Therefore it was not possible to compare the obtained results with any similar data. Although significant differences (P < 0.01) in frequency of TTMDV/SAV were observed between the healthy controls and each of the patients with HIV and HIV/HCV co-infections, no significant difference was observed between the cases with HIV and HIV/HCV co-infections. Since there was no information about the stage of clinical disease, its correlation with immunodeficiency caused by HIV remained unclear. The frequency of the virus in healthy individuals was 14.5% that was lower than frequencies reported in healthy individuals in Korea (34.5%) (8) and France (20%) (5), and higher than the reported frequency of 8.6% in Italian blood donors (6).
In the current study, the prevalence of the virus in patients with HIV and HIV/HCV co-infections was much higher than the prevalence of the virus in various diseases from other countries including acute respiratory diseases (46.7%) (1), Kawasaki syndrome (50%) (8), and cervicitis (50%) (9). Further studies are needed to determine the reasons for the high prevalence of TTMDV/SAV in the current study compared with those of the other ones, which may be due to the difference in the samples tested or the presence of some factors in the serum samples, which supports the presence and replication of the virus (possibly synergism in co-infection with two or three viruses and also the sensitivity of the assays which should be investigated).
The prevalence of the virus in the HIV positive and patients with HIV/HCV co-infections was 86% and 75% in the age group of 20 - 40 years, respectively, indicating that the individuals are most at risk of co-infection with TTMDV/SAV in this age group. It can be probably related to the same transmission route. On the other hand, co-infection with HIV/HCV may reduce the immune response and clearance of TTMDV/SAV from the body. There were not any significant differences in the means of ALT and AST levels between the patients with and without TTMDV/SAV infection in the studied groups; therefore, no association was expected between viral infection and altered levels of these enzymes. It is noteworthy that the amount of AST and ALT was higher than normal in fifty-two HIV positive individuals. Whether the virus (TTMDV/SAV) was involved in the raise of the levels of these enzymes in these cases or not, remains unknown and further investigations are required.
As shown in Figure 2, high genomic homology was observed between the obtained sequences and some of the TTMDV/SAV isolates in Korea (8). The sequences of HIV/HCV (MFa5, MFa7) and HBV/HCV/HIV (MFa1) in the co-infected patients were similar to the isolates of HCV (KR-HC-0005 ORF1, KR-HC-0023 ORF1) and HBV (KR-HB-0140 ORF1) in the co-infected individuals in Korea, also the sequence of healthy individual (MFa3) was similar to the isolate of healthy individual in Korea (SAV-KR-CT-0017 ORF1) and isolate of a cervicitis case in Iran (HQ259681 = SAV Isfahan Sal F1). Phylogenetic analysis of SAV in Korea proposed the three main groups of genotypes including G1, G2, and G3. The most prevalent genotype was G1, whose members showed high homology to SAV1 and Genotype 2 included SAV2 (20).
Comparison of the obtained sequences with the ones reported by Chung et al. (20) (MFa1, MFa2, MFa4, MFa5, and MFa6 sequences) indicated that they were placed in the same cluster with group one viruses (like SAV KR-BD-0105 Orf1 and SAV KR-BD-0078 Orf1) and MFa3 and MFa7 sequences were placed in the same cluster with group two viruses (like SAV KR-BD-0265 Orf1 and SAV KR-BD-0277 Orf1) (Figure 3). It seems that the SAV1-like viruses are more prevalent in the sampled area and consistent with the genotype distribution of SAV in Korea. Since there were only a few individuals with HIV/HCV/HBV co-infection in the current study, interpretation of their role was not possible. According to the findings of the study, the studied region is at risk of TTMDV/SAV virus. Since TTMDV/SAV was discovered a few years ago, few studies have been done and there is a poor understanding of its molecular properties and pathogenic potential.