1. Background
Urinary tract infection (UTI) involves colonization and invasion of bacteria into the tissues of the urinary tract. The infection in humans is influenced by gender and age and is common in young women (1-3). Uropathogenic Escherichia coli UPEC bacteria often originate from feces and once these microorganisms overcome the host's immune system, they are able to colonize the lower urinary tract and establish an infection (3, 4). The infection caused by UPEC is usually self-limiting and is rarely spread beyond the urethra. However, UPEC bacteria invade bladder epithelial cells by intracellular amplification, and upon exiting cell surfaces, invade other epithelial cells in the tissues of the bladder, establishing a persistent infection (2, 5). Due to urinary flow, mucus and secretory IgA secretion, and bactericidal properties of urine epithelial cells, the urinary tract is normally sterile. However, some strains of E. coli bacteria can grow in the urinary tract owing to several shared virulence factors, such as adhesion molecules and toxins (6).
The early step in the colonization of UPEC strains on mucosal surfaces of the host is dependent on adhesion molecules, such as pili S, Dr family of adhesins, pili p, and pili type I (7, 8). Among them, type I and p Pili are considered the most important (9-12). It is believed that UPEC bacteria establish UTIs and the presence of PapG possibly promotes the progression of the infection to a more severe form, called pyelonephritis (4, 9). Current treatment of acute UTI is limited due to the increasing rate of antibiotic-resistant strains as well as adverse effects of vaccines (6, 13). While several vaccine candidates have been tested in clinical trials against UTI in humans, no suitable results have been obtained to date. Therefore, the design of new strategies in vaccine development can lead to an increase in vaccine efficacy (14, 15).
A variety of studies have shown that the adsorption of vaccine candidate antigens on the surface of some bacterial cells can be used as a delivery system based on a live bacterial vector and can strongly stimulate immune responses (16, 17). A number of studies have indicated that in addition to non-pathogenic lactic acid bacteria, which act as probiotics and have significant positive effects on various aspects of the immune system, immune-modulators can also increase immune responses (15-18). Lactobacillus reuteri, a well-known probiotic, exhibits a potent immunomodulatory effect on the immune responses.
2. Objectives
In the present study, L. reuteri, was used as a host to deliver a vaccine antigen in a more immunogenic form. The vaccine candidate PapG was anchored to the surface of L. reuterivia AcmA, which was used as a linker, and this surface display system vaccine was evaluated against UTIs (17, 19). It is hypothesized that the presentation of the PapG protein on the cell surface of L. reuteri makes the vaccine more immunogenic, and thereby the immune system may be stimulated more effectively (17, 18).
3. Materials and Methods
3.1. Construction of a Recombinant PapG Expression Vector
The PapG gene segment containing a histidine tag was designed, synthesized by Bioneer, and cloned into a pEXA vector (Bioneer, Deajeon, Korea) at the NdeI/BamHI restriction enzyme sites. The recombinant pEXA vector was transformed into E. coli DH5 cells using the calcium chloride method. Then, plasmid purifications were carried out on the transformed bacteria (Bioneer). The purified plasmid was digested with NdeI and BamHI (Fermentas, Vilnius, Lithuania) and the isolated PapG gene segment was ligated into the pET21a vector. To confirm whether the pET21a vector contained the PapG gene segment, the recombinant plasmid was double digested with NdeI/ BamHI enzymes, and electrophoresis was carried out on a 1% agarose gel. Finally, the fidelity of the pET21a/PapG vector was confirmed by gene sequencing (Macrogen Institute, Seoul, South Korea).
3.2. Construction of a Recombinant PapG-AcmA Expression Vector
To bind the PapG protein to the surface of L. reuteri, the AcmA anchor protein was linked to the PapG protein. The PapG-AcmA fusion segment containing a histidine tag in the pEXA vector (at the NdeI/EcoRI sites) was purchased from Bioneer. The pEXA/PapG-AcmA vector was digested with NdeI/EcoRI enzymes and the PapG-AcmA segment was ligated into the pET21a vector. The fidelity of the pET21a/PapG-AcmA vector was confirmed using NdeI/BamHI enzymatic digestion and sequencing (Macrogen Institute).
3.3. Expression of the Recombinant PapG and PapG-AcmA Proteins
A new culture of the E. coli Origami strain was started and then recombinant pET21a/PapG-AcmA and pET21a/PapG vectors were transformed using the standard calcium chloride protocol. To express the recombinant PapG and PapG-AcmA proteins, bacteria containing each plasmid were induced for 4 hours using 1 mM IPTG. Protein expression was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant PapG and PapG-AcmA proteins were further confirmed with western blotting using anti-his antibodies.
3.4. Western Blot Analysis of the Recombinant Proteins
After the expression of PapG and PapG-AcmA in the host, SDS-PAGE was performed to separate the proteins based on the molecular weight. Subsequently, the bands were transferred to the nitrocellulose membrane in Tris-glycin buffer containing 20% methanol. The unbound membrane sites were blocked by 3% Tris-buffered saline with bovine serum albumin (TBS-BSA) containing Tween 20. The membrane was then washed three times with TBS-T, and incubated with the 6 his-tag antibody horseradish peroxidase conjugate (Roche, Mannheim, Germany) for 2 hours. Afterwards, the membrane was washed again and incubated with DAB (3, 3-diaminobenzidine) substrate solution containing 1% H2O2. When a deep brown band appeared, the membrane was washed with distilled water.
3.5. Expression and Purification of PapG and PapG-AcmA Proteins
The E. coli cells expressing the recombinant proteins were centrifuged and the cell pellet was suspended in buffer A (20 mM Tris-HCl, 0.5 M NaCl, 0.3% Triton X100, 1 mM PMSF, 10 mM imidazole, pH 8.0), and sonicated for 10 cycles of 40 seconds at 4°C (MSE, London, UK). The suspension was centrifuged (12,000 rpm, 20 minutes, 4°C), the sample was loaded on a Ni-NTA column (Qiagen, Valencia, CA, USA), and purification was performed according to the manufacturer's standard protocol. Protein refolding was carried out in urea. After dialysis of purified samples, the concentration of each protein was detected via the Bradford method.
3.6. Endotoxin Analysis of the Products
The endotoxin levels of purified PapG and PapG-AcmA were detected by the limulus amoebocyte lysate (LAL) test according to the manufacturer's standard protocol (Thermo Scientific, Waltham, MA, USA).
3.7. Surface Display of PapG-AcmA on Lactobacillus reuteri
Binding of the recombinant PapG fusion protein to the ligand on the surface of L. reuteri was carried out using the AcmA protein as an anchor. Briefly, 125 µg of PapG-AcmA in a volume of 200 µL was mixed with 3.2 × 1010Lactobacillus, incubated at 37°C for 2 hours and centrifuged at 2,000 rpm for 10 minutes. The pellet was gently washed three times in PBS to obtain a suspension that enabled PapG to be displayed at the surface of L. reuteri.
3.8. ELISA for Surface-Displayed PapG
Binding of the recombinant PapG-AcmA fusion protein to the surface of the bacteria was examined by enzyme-linked immunosorbent assay (ELISA). Briefly, ELISA plates were coated overnight at 4°C by L. reuteri displaying PapG-AcmA and untreated L. reuteri as a negative control. After coating, the plates were blocked with 3% PBS-BSA for 2 hours at 37°C. Then, 100 µL of the anti his-tag monoclonal antibody was added to each well and plates were incubated for 2 hours at 37°C. After washing 5 times, 100 µL of a 1/10,000 dilution of the anti-mouse horseradish peroxidase conjugate antibody was added for 90 minutes at 37°C. The plates were washed and 100 µL of TMB (Sigma, St. Louis, MO, USA) substrate was added to each well. The reaction was stopped after 20 minutes and the absorbance was measured at 450 nm using the ELISA Auto Reader. The ELISA was carried out on bacteria stored at 4°C for 1, 2, 3, 4, and 60 days.
4. Results
4.1. Construction of PapG and PapG-AcmA Expression Vectors
After construction of pET21a/PapG-AcmA and pET21a/PapG vectors, double digestion of recombinant plasmids was performed using NdeI/BamHI and NdeI/EcoR restriction enzymes for the pET21a/PapG-AcmA and pET21a/PapG vectors, respectively. The results of agarose gel electrophoresis of the digested plasmids showed bands at 945 bp and 629 bp for the PapG-AcmA and PapG gene segments, respectively (Figure 1). In addition, the results of sequencing confirmed both gene segments in the pET21a vector (data not shown).
A) Double digestion of the pET21a/PapG vector with NdeI/BamHI enzymes showed a 629-bp band using gel electrophoresis. Lane M, DNA marker; Lane 1, PapG gene segment. B) Enzymatic digestion of the pET21a/PapG.AcmA vector with NdeI/EcoRI enzymes showed a 945-bp band using gel electrophoresis. Lane M, DNA marker; Lane 1, PapG.AcmA gene segment.
4.2. Prokaryotic Expression of the Recombinant PapG and PapG-AcmA Proteins
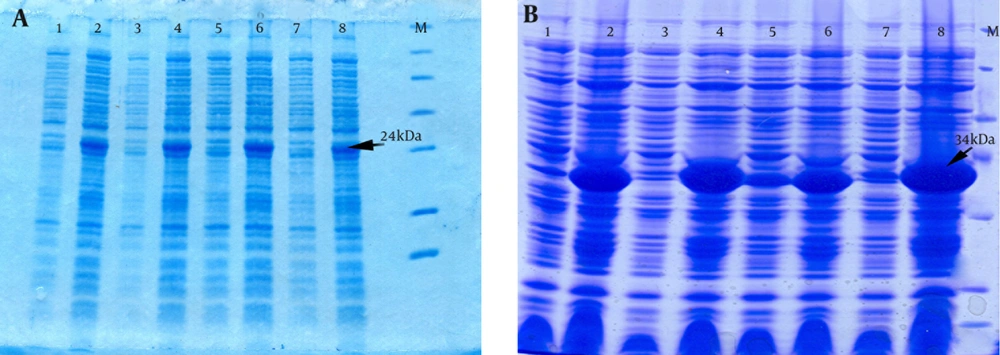
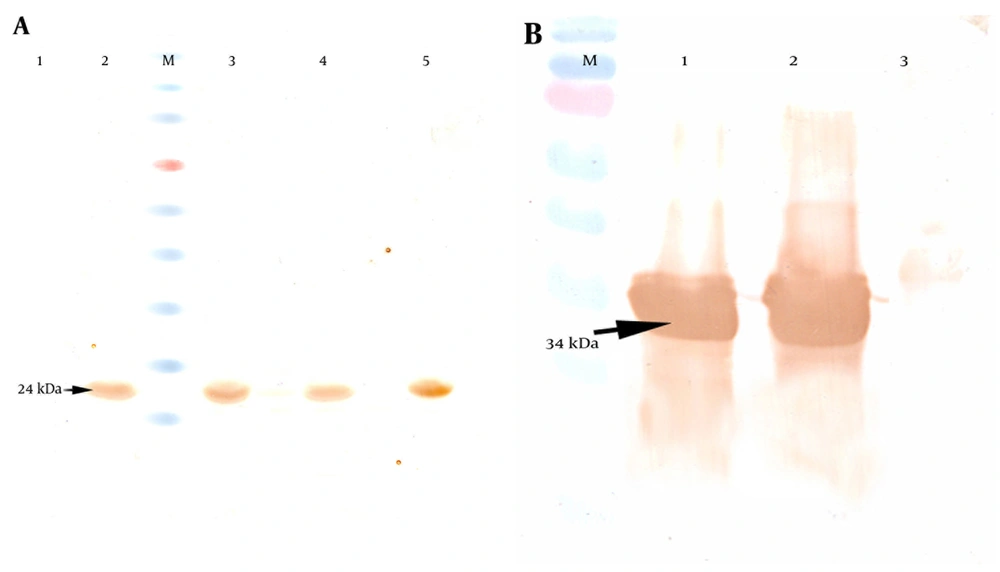
The E. coli Origami strain was used to express the recombinant PapG and PapG-AcmA proteins. After transformation and expression under 1 mM IPTG induction, SDS-PAGE indicated the overexpression of the PapG (approximately 24 kDa) and PapG-AcmA (approximately 34 kDa) proteins (Figure 2). Additionally, a western blotting analysis with the anti his-tag antibody showed bands of approximately 24 kDa and 34 kDa for the PapG and PapG-AcmA proteins, respectively (Figure 3).
A) Expression of a 24-kDa recombinant PapG with determined using SDS-PAGE in lanes 2, 4, 6, and 8 and un-induced samples are shown in lanes 1, 3, 5, and 7. M, protein size marker. B) An SDS-PAGE analysis shows a 34-kDa recombinant PapG.AcmA fusion protein in lanes 2, 4, 6, and 8 and un-induced samples in lanes 1, 3, 5, and 7. M, protein size marker.
A) Detection of the PapG protein with anti-6 his antibody revealed a 24-kDa band in western blotting. Lane 1: un-induced sample. Lanes 2, 3, 4, and 5, induced samples as negative controls; M, protein size marker. B) Analysis of the PapG.AcmA protein versus the anti-6 his antibody showed a 34-kDa band in western blotting. Lanes 1 and 2, induced samples; Lane 3 un-induced sample as a negative control; M, protein size marker.
4.3. Purification of the Recombinant PapG and PapG-AcmA Proteins
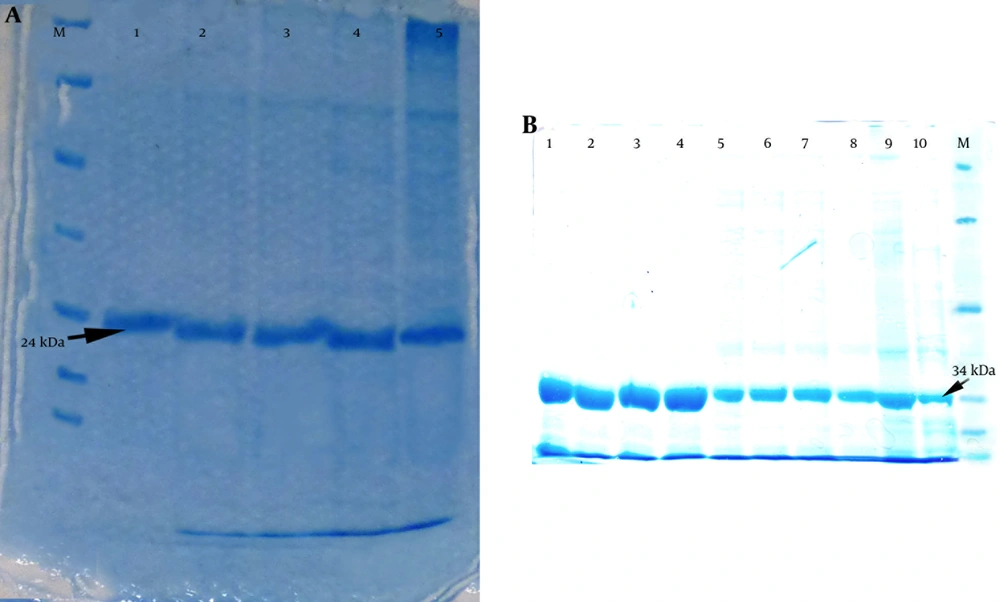
An Ni-NTA column was used to purify the recombinant proteins. After purification of the PapG and PapG-AcmA proteins and an SDS-PAGE analysis, a 24-kDa band for PapG and a 34-kDa band for PapG-AcmA were observed with minimal contamination with undesired proteins (Figure 4).
A) Purification of the recombinant PapG protein revealed a 24-kDa band using SDS-PAGE; Lanes 1 - 5 reveal different fractions of purified PapG after purification and an SDS-PAGE analysis. M: protein size marker. B) Purification of the recombinant PapG.AcmA fusion protein showed a 34-kDa band using SDS-PAGE; Lanes 1–10 show different fractions of the purified PapG.AcmA fusion protein after purification via an Ni-NTA column and SDS-PAGE; M, protein size marker.
4.4. Endotoxin Levels in the Purified Proteins
based on the LAL test, the purified recombinant PapG and PapG-AcmA proteins had endotoxin levels less than 0.07 EU/mL.
4.5. Detection of the PapG Protein on the Surface of Lactobacillus reuteri
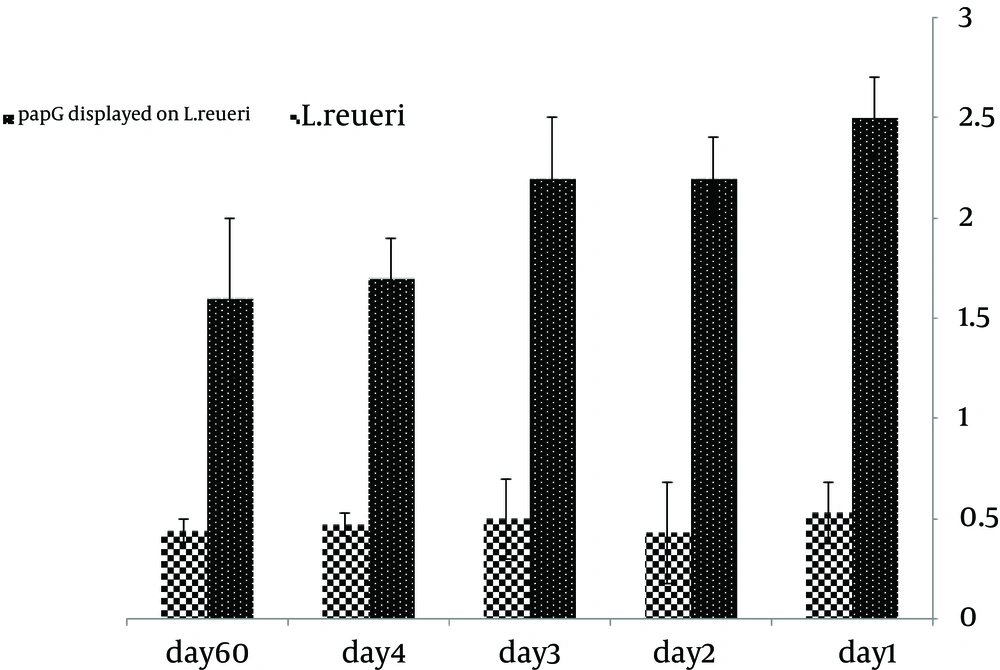
Surface display of the PapG protein was confirmed by ELISA. Considering the linkage of the PapG protein to his-tag, the antibody against the his-tag was used to detect the PapG protein on the surface of L. reuteri. The ELISA results for the surface-displayed PapG in comparison to untreated bacteria showed more significant absorbance relative to control samples. After 1, 2, 3, 4, and 60 days of storage at 4°C, an ELISA analysis of the PapG protein on the surface of the bacteria indicated a slight decrease in optical density (OD), especially after 60 days of storage, suggesting stable surface display of the PapG protein on L. reuteri (Figure 5).
5. Discussion
Escherichia coli remains one of the most common bacteria isolated from various human infections, such as UTIs, meningitis, and sepsis (1-3). Escherichia coli type P is one of the most important agents of UTIs worldwide (9-12). According to various reports, UTI occurrence is higher in women, children, the elderly, and immunocompromised patients than in men (3, 10, 12). The binding process in UPEC is facilitated by type 1 and P fimbriae (2, 9). Unlike type 1 fimbria, which mainly causes infections of the lower urinary tract system, P fimbria leads to ascending infections in the urinary tract and is the basis for the severe kidney infection pyelonephritis (2, 4, 9). Numerous studies have focused on UTIs (20-22). Antibiotics are a well-known method to reduce and prevent acute urinary infections; the development of an effective vaccine against the specific infection is another possible strategy (6, 13-15). Since the binding step is very important in the colonization and infection process, the inhibition of bacterial binding would be an effective strategy; a wide variety of vaccines focus on the inhibition of bacterial binding via distinct target molecules (13-18).
In the present study, the recombinant protein PapG, which is a critical adhesion molecule involved in the pathogenesis of bacteria, was expressed. Various studies have shown that PapG plays a critical role in the attachment and the occurrence of the infection, while inhibition of the molecule significantly inhibits pyelonephritis infections (9-12). Therefore, we hypothesized that the use of L. reuteri in combination with a vaccine modulates immune responses and increases vaccine potency. In fact, through stimulation of the innate immune system and Toll like receptors (17-19), L. reuteri provides a milieu in which lymphocytes are able to recognize PapG in inflammatory conditions and increase the immunogenic form (23-25). Furthermore, L. reuteri can act as an inhibitor of pathogenic bacteria isolated from women with bacterial vaginosis; therefore, it can have beneficial effects against UTIs (26).
In this light, the recombinant protein PapG was first produced and then confirmed using SDS-PAGE and western blotting. The immunogenic form of PapG was displayed on the surface of L. reuteri. For this purpose, the AcmA protein was used as an anchor molecule. Many studies have shown that AcmA can link to a protein on the surface of Lactobacillus spp. (17, 18). Therefore, the AcmA protein was used as a linker molecule in this study. To link the molecule to the surface of L. reuteri, the PapG protein was fused at the gene level to produce PapG-AcmA, which acted as a fusion protein for the linkage of PapG to the surface of L. reuteri. Several studies have shown that AcmA is a suitable linker for the ligation of various proteins on the surface of Lactobacillus sp. (19, 27). Herein, the PapG-AcmA fusion protein was purified and incubated with L. reuteri bacteria. The binding level of the protein to the surface of the lactobacillus cells was examined by ELISA.
The recombinant protein was displayed on the cell surfaces, suggesting that the strain can be used as a competitive inhibitor agent against UPEC in animal experiments. An ELISA analysis of the PapG protein on the surface of L. reuteri showed that the OD related to the PapG protein on the surface of L. reuteri did not decrease after 4 days of storage at 4°C, while samples stored for 60 days showed a slight decrease. Our results showed that PapG, a vaccine candidate for the surface display strategy, has a suitable stability during storage at 4°C for a minimum of one month before immunization. Several studies have confirmed the stability of the vaccine in a surface display system, consistent with our results (17-19, 27, 28).
Various studies have been conducted on the presentation of antigens at the surface of bacteria, such as Lactobacillus sp. (29, 30). The Helicobacter pylori urease subunit B gene E fragment with an anchor sequence from Staphylococcus aureus (spaX) was cloned into a PAMJ399 expression vector, expressed under a p170 promoter in L. lactis, and confirmed by western blot analysis (28). This was the first report of a surface presentation system using lactic acid bacteria to provide an oral vaccine against H. pylori (28). In 2009, Jian-kui et al. presented E. coli PgsA and F41 antigens on the surface of L. casei and the antigen display was confirmed by immunoblotting, immunofluorescence, and flow cytometry (15). In addition, oral administration of surface-displayed Lactobacillus to mice causes an increase in the secretory IgA titer and had a protective effect in an experimental challenge (15). In the present study, we produced L. reuteri bacteria displaying the PapG protein as a vaccine candidate for further study. We intend to evaluate the efficacy of this vaccine in a murine model to show the effect of our vaccine candidate on immunologic parameters as well as the protective effect against experimental challenges.