1. Background
Hepatitis E Virus (HEV) has been classified to the Orthohepevirus A species, Orthohepevirus genus and Hepeviridae family. This virus causes acute but self-limited hepatitis in the general population (1, 2). Infection with HEV ranges from moderate to severe hepatitis, yet severity of the disease increases with age (1, 2). The mortality rate can range between 25 to 31% in pregnant women, particularly during the third trimester of pregnancy (1, 3), and 30 to 70% in individuals with chronic liver disease (1, 4). No specific treatment exists for acute hepatitis E, and no protective vaccine against HEV infection has been licensed (1, 5). Therefore, development of an effective vaccine against HEV infection is the only alternative approach to prevent its spread. At present, molecular approaches are the main focus of vaccine design, since there are no appropriate and effective culture systems for HEV replication (6).
There are three overlapping open reading frames (ORF1, ORF2 and ORF3) in the genome of HEV (7). The ORF2 gene encodes capsid protein of 72 kDa, which has 660 amino acids (1, 7). The capsid protein has been studied for HEV vaccine development, as it is highly conserved and immunodominant among HEV species, and induces long-lasting immunity (6, 8). In addition, antibodies against capsid protein neutralize HEV in vitro, and protect primates against HEV infection (8, 9). This suggests that ORF2 protein could be used as a suitable antigen in serological test kits for the diagnostic and seroepidemiological surveillance of HEV infection (2, 10), and also as promising a vaccine candidate against HEV infection in humans (11).
The 112 to 660 residue of ORF2 protein self-assembles into virus-like particles (VLPs) and initiates strong immune responses (12). However, when the full-length capsid protein is expressed, its immunogenic properties are masked due to insolubility. Therefore, most efforts to develop vaccines against HEV infection have focused on the truncated or short forms of the capsid protein (5, 13). Up to now, full-length and various truncated forms (112 - 660 aa, 112 - 607 aa, 368 - 606 aa and 458 - 607 aa) of ORF2 have been used in several DNA vaccine studies (14-16). Since the time of DNA vaccine innovation, several strategies have been introduced to improve vaccine potency such as co-administration of different adjuvants, codon optimization of the gene, or using plasmids encoding a secreted form of the protein. Codon usage frequency is different among various organisms. Therefore, codon optimization can result in high expression of the desired gene similar to the host cell genes, whose expression levels are high. The mRNA secondary structures that have a negative effect on the translation efficiency are reduced or removed by codon optimization (17-19).
In vaccination with recombinant DNA, the processing and presentation of antigens are improved, since the native form of antigens are expressed in cells, and can usually activate both arms of the immune system (14, 16, 20). DNA vaccines can be formulated to target specific cell compartments for antigenic processing. However, plasmids encoding a secreted form of the target protein, by fusing it to the signal sequence of human tissue plasminogen activator (tPA), are generally more immunogenic, for both B and T cells, than plasmids encoding a non-secreted form (21, 22). A universal Pan HLA-DR (allelic DR) epitope peptide, named PADRE, has been described previously, which is capable of binding promiscuously to variants of the human major histocompatibility complex (MHC) class II molecules DR with high-affinity and is effective in both humans and mice (23). The PADRE HTL epitope was used as an adjuvant to increase the Cytotoxic T lymphocyte (CTL) responses (24). Based on this evidence, herein, we designed and constructed two DNA plasmids encoding tPAsp-PADRE-truncated ORF2 gene cassette and truncated ORF2 gene. The novel chimerical pVAX-tPAsp-PADRE-truncated ORF2 plasmid carries the secretory signal sequence derived from human tissue Plasminogen Activator (tPA) and PADRE sequence fused at the N-terminus of the truncated form of the HEV ORF2 gene.
2. Objectives
In the present study, the tPAsp-PADRE-truncated ORF2 optimized gene cassette and truncated ORF2 optimized gene from Hepatitis E Virus were cloned in the pVAX1 expression vector separately, and transfected into HEK293 cells and Chinese Hamster Ovary (CHO) eukaryotic cells. Furthermore, the main goal of the present study was to construct the recombinant plasmids pVAX-tPAsp-PADRE-truncated ORF2 and pVAX-truncated ORF2 as DNA vaccine candidates.
3. Materials and Methods
3.1. Bacterial Hosts, Plasmid, Eukaryotic Cells and Culture Conditions
Escherichia coli strain DH5α (Novagen Inc., Madison, Wis., USA) was used for transformation and the pVAX1 plasmid (Invitrogen®, Carls-bad, CA, USA) was used for subcloning of the truncated ORF2 and tPAsp-PADRE-truncated ORF2 (112 - 660 aa) gene cassette. Eukaryotic expression vector, pVAX1, was constructed to be consistent with the Food and Drug Administration (FDA) regulations. HEK293 cells, routinely maintained in our laboratory, and CHO cell line, purchased from the national cell bank of Iran (Pasteur Institute), were used for protein expression. All kits were purchased from the Qiagen company (Germany); T4 DNA ligase and restriction endonucleases were purchased from New England Biolabs (New England Biolabs Inc., USA). All chemicals were purchased from Merck (Germany) and Sigma-Aldrich Corporation (Germany).
3.2. Design, Optimization and in Silico Analysis of tPAsp-PADRE-Truncated ORF2 Gene Cassette of Hepatitis E Virus and de novo Synthesis
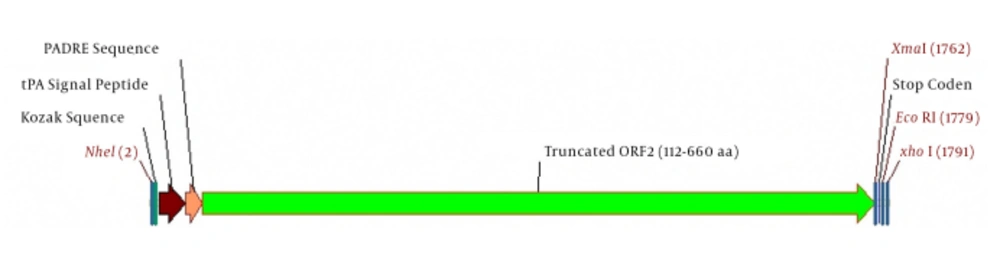
The 112 - 660 amino acid sequence of ORF2 gene of Hepatitis E virus genotype 1 (Sar55 strain) was retrieved from UniProtKB/Swiss-Prot databases (Pakistan/Sar-55 strain, accession number P33426) and entered in the Vector NTI software (Vector NTI software version Advance® 11.5, Invitrogen). Next, the following sequences were added to the N-terminal of the truncated ORF2 (aa 112-660); primarily, the 22-amino-acid-sequence (MDAMKRGLCCVLLLCGAVFVSP) of the secretory peptide of human tissue-type plasminogen activator protein, (tPAsp signal peptide) was retrieved from http://www.signalpeptide.de/ and UniProtKB/Swiss-Prot databases (Accession no. P00750). In the next step, PADRE 13-amino acid sequence (Pan HLA DR-reaction epitope) that is the universal epitope of T lymphocyte (CD4+ T cell epitope), AKFVAAWTLKAAA, was placed after the secretory peptide sequence. All Sequences were back translated to sequence of nucleotides using Vector NTI (Vector NTI software version Advance® 11.5, Invitrogen). The reverse translation used the most abundant codon for each amino acid, found in a compendium of highly expressed mouse genes. Finally, the complete kozak sequence, GCCGCCACCATGG, was added to increase the efficiency of translation initiation. Two restriction enzyme digestion sites, including NheI and XhoI, were placed in the tPAsp-PADRE-truncated ORF2 gene cassette for sub-cloning into the pVAX1 vector. Arrangement of fragment junctions is shown in Figure 1. Also to ensure the accuracy of the cloning process, virtual cloning was performed using the clone Manager software (Clone Manager Professional 9, USA) in pVAX1 eukaryotic expression plasmid.
To optimize the gene cassette, the in silico analysis was performed using online data bases such as codon adaptation index calculation (http://genomes.urv.es/CAIcal/), Analysis of Codon Usage (http://codonw.sourceforge.net), Codon Database (http://www.kazusa.or.jp/codon), Codon-Adaptation Tool (http://www.jcat.de/), signal peptide analysis (http://www.cbs.dtu.dk/services/SignalP/), and Leto (Entelechon, Germany). The gene cassette was then analysis using GenScript rare codon analysis the tool software (www.genscript.com) was used for expression in mouse cells and submitted to GenScript for more optimization and verification using the GenScript’s OptimumGene gene design tool (GenScript Inc., NJ, USA). This software optimizes a variety of parameters that are critical for efficiency of gene expression such as codon usage, eliminates immature polyadenylation sites, splicing sites, killer motifs, repeated sequences and RNA secondary structure, and also avoids excessive local content of GC or AT. Following verification of the construct’s properties, the optimized tPAsp-PADRE-truncated ORF2 gene cassette was then synthesized and cloned into the pBMH commercial cloning vector by Biomatik Company (Biomatik Corporation, Cambridge, Canada).
3.3. Sub-Cloning of Codon-Optimized tPAsp-PADRE-Truncated ORF2 Gene Cassette in pVAX1 Eukaryotic Expression Vector
To sub-clone the optimized tPAsp-PADRE-truncated ORF2 gene cassette into pVAX1 eukaryotic expression vector, the recombinant plasmid pBMH-tPAsp-PADRE-truncated ORF2 (aa 112-660) and the pVAX1 plasmid were separately digested by NheI and XhoI restriction enzymes. After analysis on agarose gel, the linearized pVAX1 plasmid and the tPAsp-PADRE-truncated ORF2 (aa 112 - 660) gene were extracted from 1% agarose gel using the QIAquick® Gel Extraction Kit (Qiagen, Germany) and ligated by T4 DNA ligase (New England Biolabs Inc., USA). The recombinant plasmid pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660) was constructed and transformed in to Escherichia coli DH5α competent cells by electroporation, and selected on Luria-Bertani agar containing kanamycin, as described previously (25). Several colonies were evaluated by the colony PCR method and restriction digestion with NheI and XhoI enzymes. Next, the recombinant plasmids with the correct restriction pattern were used for DNA sequencing with the T7 forward and BGH reverse primers (Bioneer, Korea), to corroborate proper orientation of the cloned gene cassette. The confirmed construct was named pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660).
3.4. Generation of a Codon-optimized Truncated ORF2 Gene and Cloning in pVAX1 Eukaryotic Expression Vector
The gene fragment encoding truncated ORF2 (aa 112 - 660) gene (without tPA secretory signal and PADRE sequence) was PCR amplified using Q5 High-Fidelity DNA Polymerase (New England BioLabs, USA). Sequence of the forward primer, which includes a Kozak consensus sequence, was 5’-AACTAGGCTAGCCGCCACCATGGCGGTGGCCCCCGCTCACGACA-3’, and sequence of the reverse primer was 5’-ACGGGCCCTCTAGACTCGAGTCATCAGAATTCTCA-3’. This gene was directly cloned into pVAX1, using restriction sites NheI and XhoI. After analysis of the truncated ORF2 (aa 112 - 660) PCR product by agarose gel electrophoresis, the specific band was extracted from 1% agarose gel using the QIAquick® Gel Extraction Kit (Qiagen, Germany), according to the manufacturer’s instructions. The purified PCR product and the pVAX1 plasmid were digested by NheI and XhoI restriction enzymes, extracted from LMP agarose gel, and purified fragments were used for ligation by T4 DNA ligase. The recombinant plasmids were electroporated into E. coli DH5α cells. Cloning of truncated ORF2 gene in pVAX1 was verified using enzyme digestion, colony PCR by universal primers (T7, BGH) and finally sequencing. The confirmed construct was named pVAX-truncated ORF2 (aa 112 - 660).
3.5. Expression of Recombinant tPAsp-PADRE-Truncated ORF2 and Truncated ORF2 in Mammalian Cells
The recombinant plasmids pVAX-tPAsp-PADRE-truncated ORF2 and pVAX-truncated ORF2, were extracted by the Plasmid Purification Midi Kit with transfection grade according to the manufacturer’s instructions (Qiagen, Germany). To evaluate expression of recombinant proteins in eukaryotic cells, ultrapure expression plasmids containing truncated ORF2 were transfected in HEK-293 and CHO cells by X-tremeGENE 9 DNA transfection reagent (Roche, Germany), according to the manufacturer’s instructions. Furthermore, HEK-293 and CHO cells as expression organisms were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (GIBCO-BRL) supplemented with 10% fetal bovine serum (FBS), in the presence of gentamycin (50 µg/mL) and amphotericin B (2.5 µg/mL) (GIBCO-BRL) at 37°C with 5% CO2, then transiently transfected with either recombinant plasmids or pVAX1 in six-well culture plates (Nunc, Denmark). After 48 hours, mRNA of truncated ORF2 gene was determined by RT-PCR in transfected cells. In brief, total cellular RNA was extracted by the RNeasy Mini Kit (Qiagen, Germany), and RT-PCR was then performed using a QIAGEN One Step RT-PCR Kit (Qiagen, Germany), according to the manufacturer’s protocol by specific primers (upstream primer: F1478 5’- ACCACAGCCAGTGATCAGC-3’ and downstream primer: R1730 5’-TCATCTTCAGCCTCTGCAG-3’).
The RT-PCR product was then electrophoresed on 2% agarose gel. The expression of recombinant truncated ORF2 protein in transfected CHO and HEK293 cells was also determined by the indirect immunofluorescence test using an Fluorescein Isothiocyanate (FITC) -labeled antibody, as described previously (26). Briefly, the transfected cells were fixed with cold methanol–acetone (1:1) for 20 minutes, and then rinsed and blocked with 2 mL of 0.15 M Phosphate Buffered Saline (PBS) containing 1% Bovine serum albumin (BSA). The cells were incubated with polyclonal anti-HEV ORF2 antibody, diluted 1:100 in PBS for 60 minutes at room temperature. The cells were rinsed again and stained with 10 µg/mL FITC-labeled anti-mouse IgG (Biolegend, USA). Next, 1 µg/mL of 4’, 6-diamidine-2’-phenylindole dihydrochloride (DAPI) (Roche, Germany) was used for staining the nuclei of the cells. After rinsing with PBS, cover slips were immediately visualized under a fluorescence microscope (Nikon, USA).
4. Results
4.1. Bioinformatics Analysis of tPAsp-PADRE-Truncated ORF2 Gene
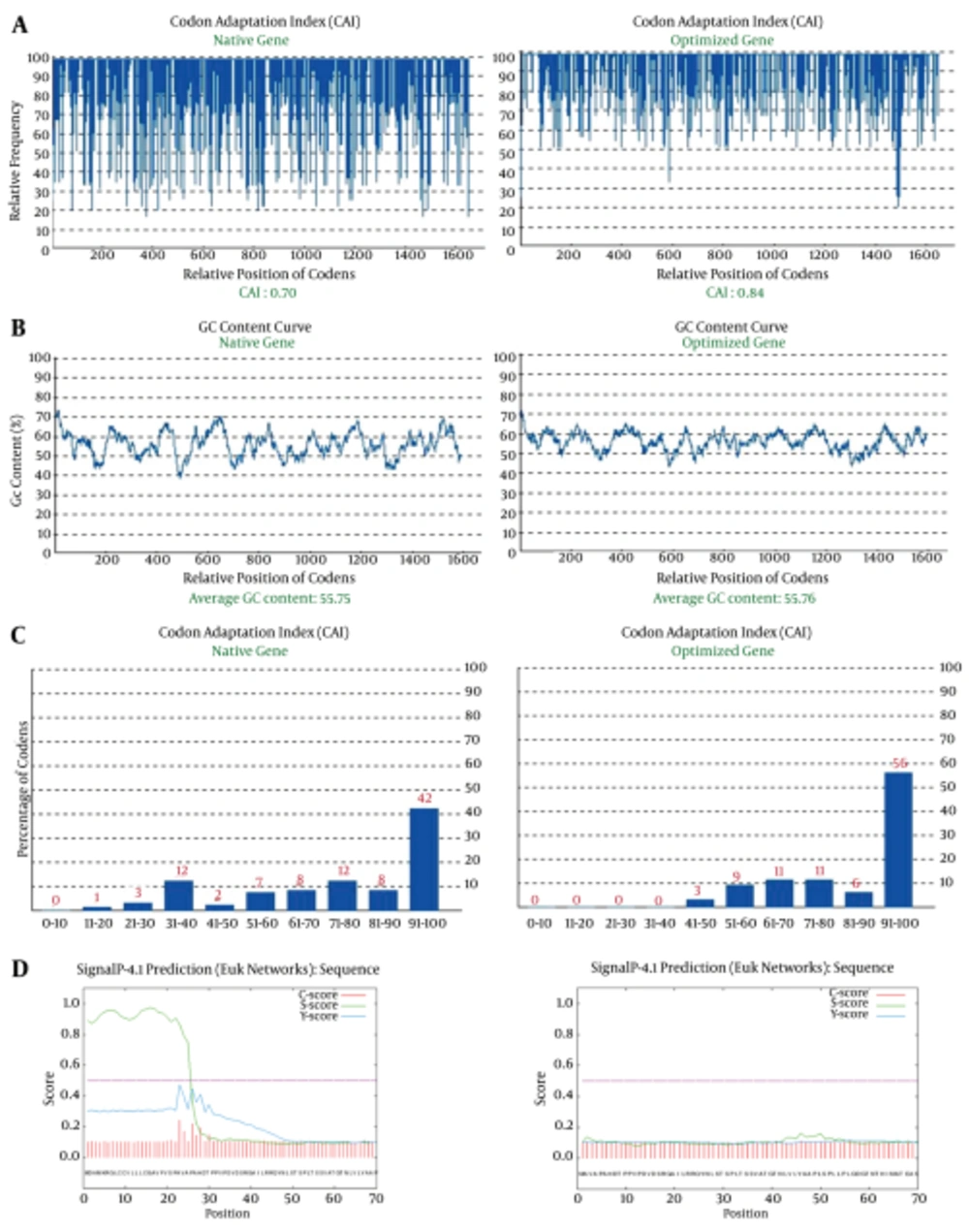
The gene cassette was optimized based on mouse favorite codons (preferred codons, which found in genes with high expression leve) using GenScript’s Optimum Gene Design (www.genscript.com). A total of 425 nucleotides were replaced to optimize codon usage for mouse cells by changing individual nucleotides without altering the amino acid sequence. Protein expression level in the desired expression system is associated with the value of CAI (27). A CAI of 1.0 is excellent while > 0.8 is good. The CAI of non-optimized and optimized genes was 0.7 and 0.84, respectively (Figure 2A). In addition to codon modifications, one splice site (nucleotide sequence GGTAAG at position 543 - 544 amino acid) was modified, and the undesirable peak with high GC content was removed (Figure 2B). Frequency of optimal codons (FOP) value of the non-optimized truncated ORF2 gene was 42%, and after optimization FOP value was improved to 56% (Figure 2C). Frequency of optimal codon values lower than 30, decrease the expression efficiency. The accuracy and performance of the secretory signal was evaluated before and after addition to sequence truncated ORF2 by using online software (http://www.cbs.dtu.dk/services/SignalP/). The results of the evaluation are shown in Figure 2D.
A, Codon Adaptation Index of the non-optimized and optimized truncated ORF2 gene; B, GC content of the non optimized and optimized truncated ORF2 gene; C, Frequency of optimal codons (FOP) of the non optimized and optimized truncated ORF2 gene; D, Presence (left picture) and absence (right picture) of 22-amino acid secretory peptide before and after addition to truncated ORF2 protein using the signal peptide analysis online software.
4.2. Sub-cloning Results of Optimized tPAsp-PADRE-Truncated ORF2 (aa 112-660) Gene Cassette in the pVAX1 Eukaryotic Plasmid
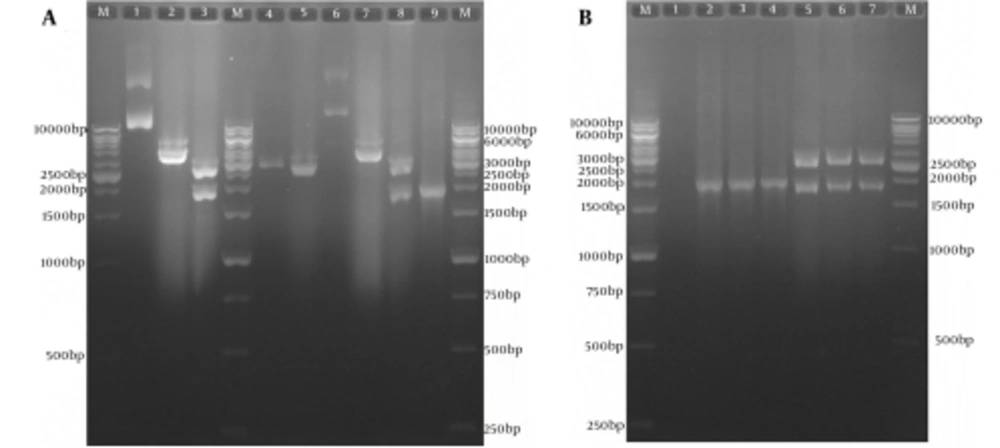
The subcloning was confirmed by colony PCR and restriction enzyme digestion (Figure 3A and B). In order to confirm the sequence of the optimized gene in the expression recombinant plasmid pVAX-tPAsp-PADRE-truncated ORF2, sequencing of both strands with T7 forward and Bovine Growth Hormone (BGH) reverse primers was carried out (Bioneer, Korea). The result of the sequence analysis of the optimized gene using Mega4 software (Biodesign Institute, Tempe, AZ, USA) showed 100% concordance with the nucleotide sequence of the gene cassette in the initial recombinant pBMH-tPAsp-PADRE-truncated ORF2.
A, Sub-cloning Results of Optimized tPAsp-PADRE-Truncated ORF2 (aa 112 - 660) Gene Cassette In Pvax1 Eukaryotic Plasmid; Lane M, 1kb DNA marker; Lane 1, pBMH-tPAsp-PADRE-truncated ORF2 plasmid; Lane 2, digested pBMH-tPAsp-PADRE-truncated ORF2 plasmid by NheI restriction enzyme; Lane 3, digested pBMH-tPAsp-PADRE-truncated ORF2 plasmid by NheI and XhoI restriction enzymes and production of two expected fragments, a 2900-bp and a 1795-bp fragment; Lane 4, pVAX plasmid; Lane 5, digested pVAX plasmid by NheI and XhoI restriction enzymes and production of 2909-bp band; Lane 6, pVAX-tPAsp-PADRE-truncated ORF2 plasmid; Lane 7, digested pVAX-tPAsp-PADRE-truncated ORF2 plasmid by the NheI restriction enzyme; Lane 8, digested pVAX-tPAsp-PADRE-truncated ORF2 plasmid by NheI and XhoI restriction enzymes and production of two expected fragments, a 2909-bp and a 1795-bp fragment; Lane 9, 1876-bp band in colony PCR test with T7 and BHG universal primers; B, Restriction Enzyme analyses and colony polymerase chain reaction results of three colonies containing pVAX-tPAsp-PADRE-truncated ORF2 plasmid; Lane M, 1 kb DNA marker; Lane 1, negative control; Lanes 2 - 4, 1876-bp band in colony PCR test; Lanes 5 - 7, digested pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660)-linker plasmid by NheI and XhoI restriction enzymes and production of two expected fragments, a 2909-bp and a 1795-bp fragment
4.3. Cloning of Codon-Optimized Truncated ORF2 (aa 112 - 660) Segment in pVAX1 Eukaryotic Expression Vector
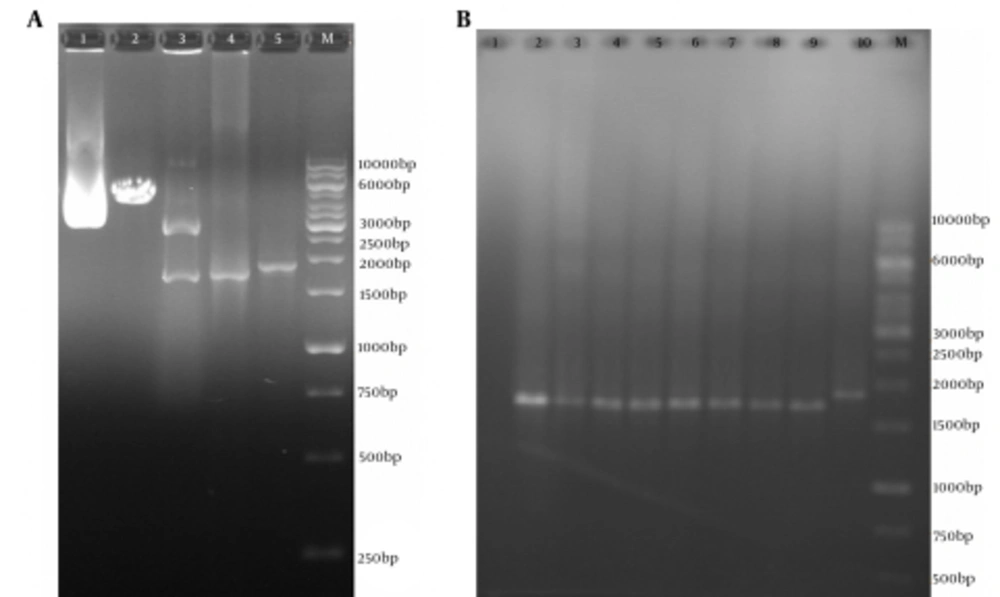
After cloning of the truncated ORF2 PCR product (without PADRE and tPAsp sequences) in the pVAX1 vector, the recombinant plasmids were detected by colony PCR and restriction enzyme digestion with NheI and XhoI enzymes (New England BioLabs, USA) (Figure 4). DNA sequencing with the T7p and BGH primers (Bioneer, Korea) confirmed that the truncated ORF2 sequence had been cloned in the proper orientation and was 100% identical to the primary codon-optimized gene.
A, Polymerase Chain Reaction Results of pVAX-Truncated ORF2 (aa 112 - 660) Expression Plasmid (without PADRE and tPAsp sequences); Lane M, 1 kb DNA marker; Lane 1, pVAX-truncated ORF2 (aa 112 - 660) plasmid; Lane 2, digested pVAX-truncated ORF2 (aa 112 - 660) plasmid by NheI restriction enzyme (4596 bp fragment); Lanes 3, digested pVAX-truncated ORF2 (112 - 660) plasmid by NheI and XhoI restriction enzymes and production of two expected fragments, a 2909-bp and a 1693-bp fragment; Lanes 4, PCR product of truncated ORF2 (aa 112-660) gene (without PADRE and tPAsp sequences) with specific primers (1710 bp); Lanes 4, PCR product of tPAsp-PADRE-ORF2 (aa 112 - 660) gene (with PADRE and tPAsp sequences) with T7p and BGH primers (1876 bp); B, Colony Polymerase Chain Reaction Results of Colonies Containing the pVAX-Truncated ORF2 (aa 112 - 660) Expression Recombinant Plasmid; Lane M, 1 kb DNA marker; Lane 1, negative control; Lanes 2 - 9, 1774 bp band of pVAX-truncated ORF2 (aa 112 - 660) plasmid (without PADRE and tPAsp sequences) due to colony PCR assay with T7p and BGH universal primers; Lane 2, 1876 bp band of pVAX-tPAsp-PADRE-ORF2 (aa 112 - 660) plasmid (with PADRE and tPAsp sequences) due to colony PCR assay with T7p and BGH primers as controls
4.4. Expression and Confirmation of the Recombinant tPAsp-PADRE-Truncated ORF2 and Truncated ORF2 in Mammalian Cells
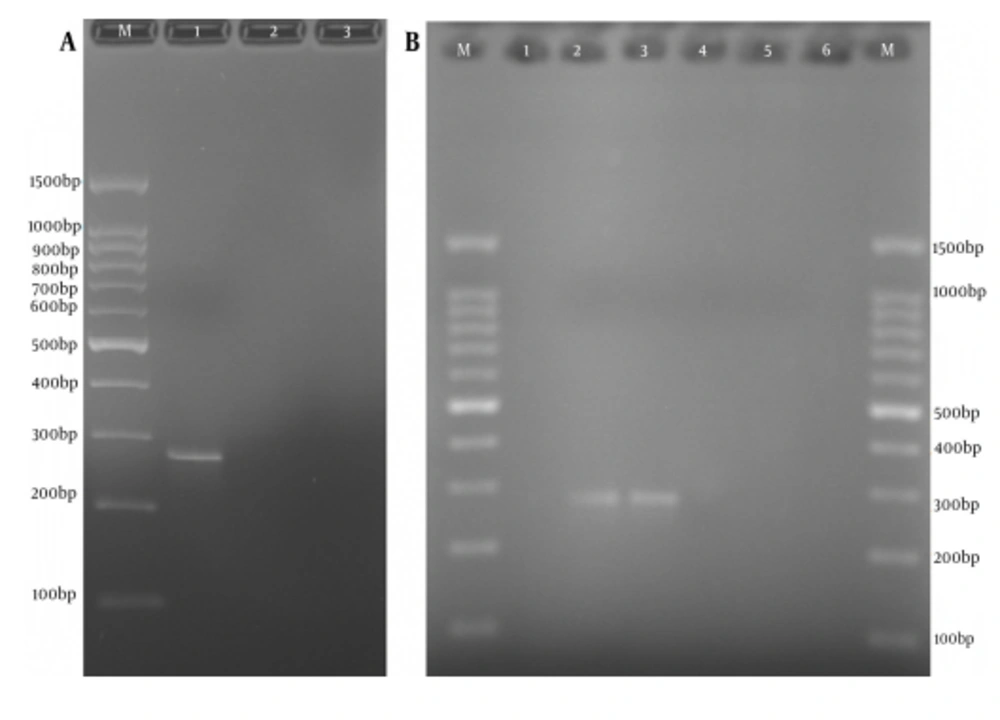
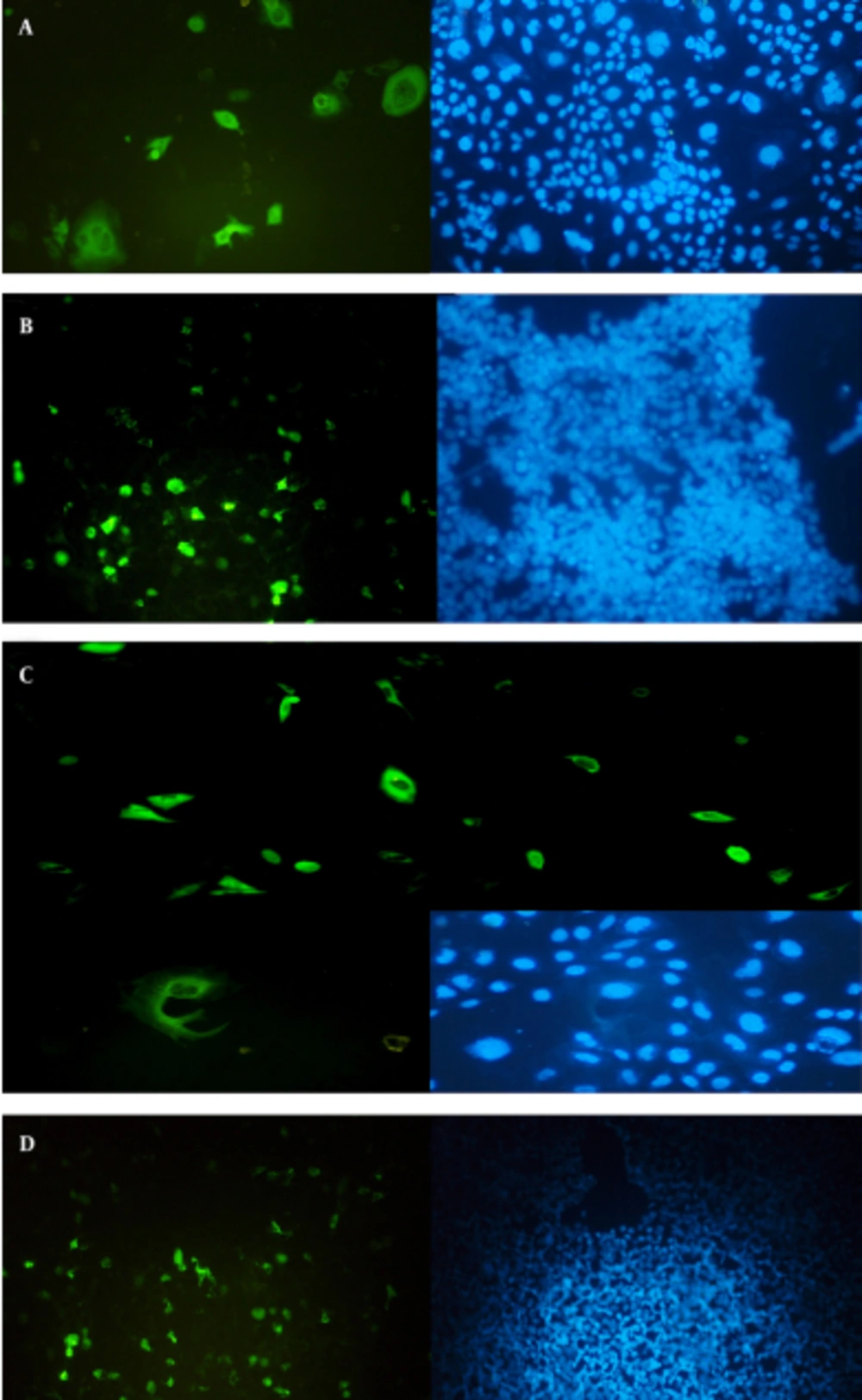
Confirmation of the truncated ORF2 gene expression in eukaryotic cells was performed at transcription and translation levels. The results of the RT-PCR assay that verify the transcription of HEV truncated ORF2 gene mRNA in transfected cells, with expression plasmids pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660) and pVAX-truncated ORF2 (aa 112 - 660), are shown in Figure 5. The size of the PCR product was 271 bp. Forty-eight hours after transfection, expression and production of truncated ORF2 protein in transfected cells was evaluated by Indirect Immunofluorescence Test (IFAT) using anti HEV polyclonal antibody. The results of the Immunofluorescence Test in transfected CHO and HEK293 cells with pVAX-tPAsp-PADRE -truncated ORF2 (aa 112 - 660), pVAX-truncated ORF2 (aa 112 - 660) and pVAX plasmid (as negative control) are shown in Figure 6.
A,Reverse Transcription-Polymerase Chain Reaction Amplification of Truncated ORF2 (aa 112-660) Gene From Transfected Cells and Electrophoresis on 2% Agarose Gel; Lane M, 100 bp DNA marker; Lane 1, RT-PCR product of truncated ORF2 (112 - 660) gene by F1478 and R1730 specific primers with 271-bp length, from CHO cells transfected with expression recombinant plasmid pVAX-truncated ORF2 (aa 112 - 660); Lane 2, RT-PCR assay of truncated ORF2 (112 - 660) gene from CHO cells transfected with pVAX1 plasmid (negative control); Lane 3, RT-PCR assay of truncated ORF2 (112 - 660) gene from untransfected cells (negative control); B, Reverse transcription-polymerase chain reaction amplification of tPAsp-PADRE-Truncated ORF2 (aa 112 - 660) Gene From Transfected Cells and Electrophoresis on 2% Agarose Gel; Lane M, 100 bp DNA marker; Lane 1 and 6, RT-PCR assay of truncated ORF2 gene from untransfected cells (negative control); Lanes 2 and 3, RT-PCR product of truncated ORF2 (aa 112 - 660) gene by F1478 and R1730 specific primers with 271-bp length, from CHO and HEK293 cells transfected with expression recombinant plasmid pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660); Lanes 4 and 5, RT-PCR assay of truncated ORF2 gene from cells transfected with the pVAX plasmid (negative control).
A, Detection of HEV truncated ORF2 protein in CHO cells and B, HEK293 cells transfected with pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660) recombinant plasmid by the IFAT test. Detection of HEV ORF2 protein in CHO cells and C, HEK293 cells; D, transfected with pVAX-truncated ORF2 (aa 112 - 660) recombinant plasmid by the IFAT test.
5. Discussion
One strategy to enhance DNA vaccine potency is codon optimization, which is a generic technique to increase the efficiency of DNA expression vectors used in DNA vaccination, to achieve optimum expression of a foreign gene in the host’s cell system (28). Bradel-Tretheway et al. showed that gene codon-optimization is associated with a profound (10 - 100 fold) increase in protein expression in human (293A, HSG, K562) or hamster (CHO) cell lines (29). He et al. used Cos-7 cells to confirm the expression of the pJHEV ORF2 plasmid (14). In the present study, we evaluated the ORF2 expression in CHO and HEK293 cells by the IFA test. The results showed that the expression level of the recombinant plasmids was higher in transfected CHO cells than in HEK293 cells, yet a greater number of HEK293 cells was transfected. The results by several groups have clearly demonstrated that vaccine vectors exploiting codon-optimized genes elicit higher levels of protein expression and humoral and cellular responses than DNA vaccine constructs encoding the corresponding wild-type gene (30-33).
Several reports and web-based programs have proposed algorithms for gene optimization on the basis of different calculation methods (34-36). In this study, the Vector NTI software was used for designing of the construct. Initially, a kozak sequence, a tissue plasminogen activator signal sequence and PADRE sequences were added to the N-terminal of the truncated ORF2 (112 - 660 aa), respectively. Subsequently, the construct was optimized based on mouse codon usage using the GenScript software to improve the expression of a truncated ORF2 DNA vaccine in mammalian cells. In wild-type ORF2 (112 - 660 aa), the Arg, Ser and Pro amino acid residues are encoded by codons rarely seen in mouse genes. By substituting high-usage codons for these rare codons without altering the amino acid sequences, we could increase the level of protein expression in mammalian cells.
Codon usage preference in a gene is determined by the codon adaptation index (CAI) and scores from zero to one (27). The designed genes with maximum CAI have high protein expression level (37). Narum et al. showed that increasing the rate of the CAI in an optimized gene plays an important role in enhancing gene expression (31). In this study, the score of CAI after codon optimization was 0.84, thus this is indicative of better chances of high-level protein expression. Any peak outside the 45% to 65% range of GC content will adversely affect transcriptional and translational efficiency, thus undesirable picks with high GC content were removed (Figure 2). Moreover, the negative CIS element with the nucleotide sequence GGTAAG in 543 to 544 amino acid positions (splice site) of non-optimized ORF2 sequence was modified during the optimization process. Negative CIS elements are motifs that negatively regulate gene expression at the level of transcription and translation.
Another indicator of codon usage index that was introduced by Ikemura is frequency of optimal codon usage (Fop), which is also used to predict the level of expression (38). The FOP value of 100 shows the highest usage frequency for a certain amino acid in a specific expression organism. After analyzing the gene sequence of truncated ORF2, we found that almost 15% of codons showed a value less than 40 for frequency of occurrence in mouse genes. Such codons are associated with low levels of protein expression because of ribosome stalling and failed translation. After optimization, 56% were calculated to have a FOP value ranging from 91 to 100 (Figure 2).
Several reports indicate that the efficiency and magnitude of immune responses elicited by DNA vaccines are influenced by the secretion ability of the expressed antigen (39, 40). Secretion of the protein may increase antigen presentation in association with MHC class II, and thus increase the activation of antigen-specific CD4+ T-cells. Transient-transfection experiments have demonstrated that plasmids encoding a secreted form of the protein compared with plasmids encoding the non-secreted form have higher expression levels, and this is probably the most important factor involved in the enhancement of immunogenicity (22). The effect of the tPA leader sequence is outstanding for weakly immunogenic plasmids or when DNA is used at suboptimal doses or at a low dose number (21). Therefore, in this study the tissue Plasminogen Activator (tPA) leader sequence was placed upstream of PADRE-truncated ORF2 sequences to direct translation products to the secretory pathway, and to target the truncated ORF2 to the Class II processing pathway.
One strategy to enhance potency of a DNA-based vaccine is to induce CD4+ T helper cell immune responses. CD4+ T helper cells are known to play an important role in the generation of CD8+ T cell immune responses as well as memory T cell responses (41). Thus, design of an immunization regimen that induces CD4+ T cell immune responses is desirable. PADRE peptide can bind to variants of the human MHC class II molecules DR with high-affinity and induce helper-facilitated immunity in mice and lymphoproliferation in human peripheral blood mononuclear cell (PBMC) (23). Pan DR T helper epitopes (PADRE) peptide has been used in conjunction with different forms of vaccines to enhance the potency of vaccines in preclinical models (42, 43). Previous studies have shown that PADRE could be used as an adjuvant in prophylaxis and therapeutic vaccines. Therefore in the present study, similar to the study of Li et al., the PADRE sequence was designed for the N-terminal of truncated ORF2 (aa 112 - 660) (43).
Overall, results of this study showed that pVAX-tPAsp-PADRE-truncated ORF2 (aa 112 - 660) and pVAX-truncated ORF2 (aa 112 - 660) recombinant plasmids encoding the truncated form of HEV capsid protein were successfully expressed in eukaryotic cells. However, the immunogenicity and potency of these recombinant plasmids require to be evaluated in vivo as two novel DNA vaccines in future investigations.