1. Background
Emergence and spread of multidrug-resistant (MDR) Acinetobacter baumannii in nosocomial settings has become a serious concern in clinical practice. The organism commonly targets critically ill patients in intensive care units (ICU) and burn wards (1). The infections caused by this organism include bacteremia, ventilator-associated pneumonia, meningitis, and infections of the urinary tract, wounds, and burns (1, 2). Acinetobacter baumannii exhibits a variety of resistance mechanisms to all existing antibiotic classes, including active drug efflux, modification of drug target sites, and most importantly, enzymatic inactivation of the antimicrobial agents (2, 3). Carbapenems, especially imipenem, is often used as the drug of choice for treatment of A. baumannii infections. However recently, high levels of resistance have emerged, causing treatment failure (4, 5). The most common mechanism responsible for carbapenem resistance in A. baumannii is production of carbapenemases, including metallo-β-lactamases (MBLs) and class D β-lactamases (oxacillinases) which, class D β-lactamases have been reported as the most widespread carbapenemases in A. baumannii isolates (2, 5-8).
The first oxacillinase found in A. baumannii was observed in an imipenem-resistant strain in Scotland in 1985, which was later designated as OXA-23. Today, over 220 OXA-type enzymes have been reported (9). Among these, five main phylogenetic subgroups have been recognized in imipenem-resistant A. baumannii, including OXA-23-, OXA-24/40-, OXA-51-, OXA-58-, and more recently, OXA-143-like oxacillinases (10). The largest subgroup is the chromosomally encoded OXA-51, followed by OXA-23-like enzymes (10, 11). However, plasmid-mediated OXA-51 genes have also been reported (12). blaOXA-23- and blaOXA-24/40-like genes can be chromosomal or plasmid-mediated, whereas blaOXA-58-like genes are mostly plasmid-mediated (11). Rapid development of MDR strains, mostly owing to the horizontal transfer of antibiotic determinants among the isolates, makes it imperative to identify the underlying resistance mechanisms and study the epidemiology of these infections. Among the fingerprinting methods used, randomly amplified polymorphic DNA (RAPD) has been widely used to detect polymorphisms in many organisms, including A. baumannii.
2. Objectives
The objective of this study was to compare the presence of blaOXA-23-, blaOXA-24/40-, blaOXA-51-, and blaOXA-58-like genes in two groups of burn and non-burn clinical isolates of A. baumannii and study the clonal spread and heterogeneity of the isolates.
3. Materials and Methods
3.1. Bacterial Isolates
Fifty-eight A. baumannii isolates were collected from Imam Hossein (30 non-burn) and Shahid Motahari (28 burn) Hospitals from October 2011 to April 2012 in Tehran. Table 1 shows the distribution of different specimen types among burn and non-burn clinical isolates of A. baumannii. Phenotypic identification of the isolates was confirmed by standard biochemical tests; the isolates were maintained in brain heart infusion broth (Oxoid, UK) containing 10% dimethyl sulfoxide (v/v) at -20°C.
| Isolate Group | Number of Isolates in Each Specimen Type | |||||
|---|---|---|---|---|---|---|
| Wound | Sputum | Blood | Catheter | CSF | Trachea | |
| Burn | 25 | 3 | 0 | 0 | 0 | 0 |
| Non-burn | 3 | 16 | 5 | 3 | 2 | 1 |
a Abbreviation: CSF, cerebral spinal fluid.
3.2. Imipenem Susceptibility Testing
The MIC and MBC of imipenem (Moon Chemicals, China) were determined by broth microdilution, according to the guidelines recommended by the Clinical and Laboratory Standards Institute (13).
3.3. Detection of OXA-Like Genes by PCR
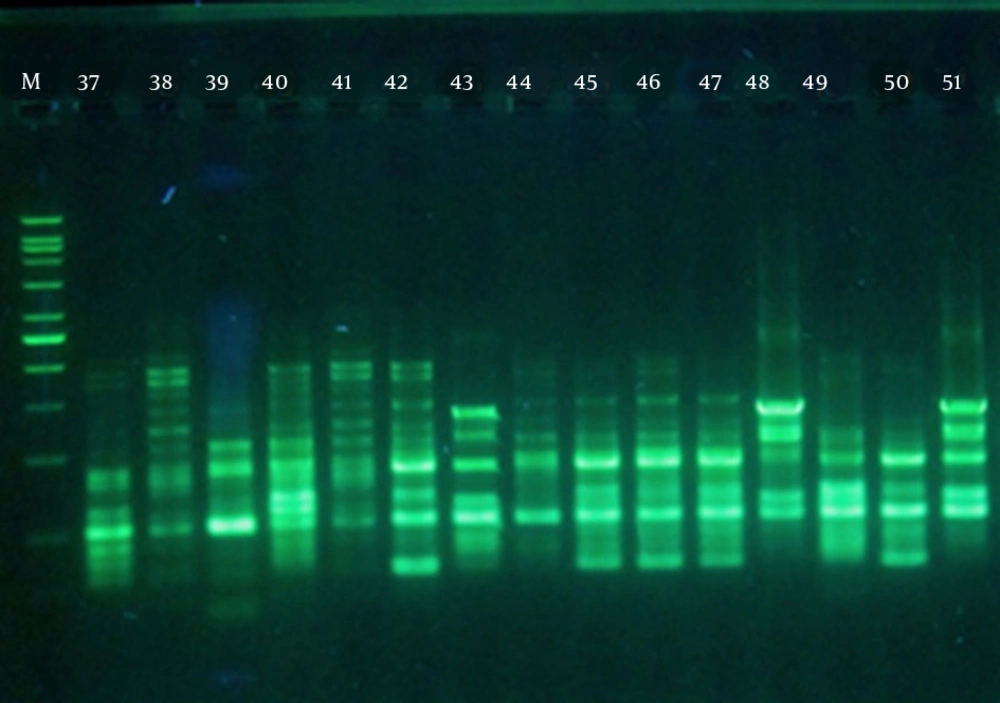
DNA extraction was carried out by boiling. Briefly, a loopful of an overnight grown culture of each test isolate was suspended in 500 μL of sterile double-distilled water, boiled at 100°C for 10 min, and centrifuged at 13,000 × g for 10 minutes. The supernatant was then used as the DNA template for PCR amplification. blaOXA-23-, blaOXA-24-, blaOXA-51-, and blaOXA-58-like genes were detected by PCR using the primers listed in Table 2 (14). PCR reaction mixtures (25 μL) contained 1 μL of DNA template, 1.4 mM MgCl2, 0.2 mM each dNTP, 0.4 μM primer, and 0.625 U of Taq DNA polymerase in the buffer provided by the manufacturer (CinnaGen, Iran). The amplifications were performed in a thermocycler (Techne TC-3000G; UK), with the following program: initial denaturation at 94°C for 5 minutes, 33 cycles of 94°C for 25 seconds, 53°C for 40 seconds, and 72°C for 50 seconds, followed by a final elongation step at 72°C for 6 minutes (11). A. baumannii NCTC 13304 harboring blaOXA-23 and A. baumannii NCTC 12156 carrying blaOXA-51 were used as controls (15). PCR products were separated on 1% agarose gels and observed after staining with RedSafe (iNtRON Biotechnology, Korea) using an image analysis system (UVLtec; St John's Innovation Centre, UK).
| Primer | Sequence | Product Size, bp |
|---|---|---|
| OXA-23 | 501 | |
| Forward | 5´-GATCGGATTGGAGAACCAGA-3´ | |
| Reverse | 5´-ATTTCTGACCGCATTTCCAT-3´ | |
| OXA-24 | 246 | |
| Forward | 5´-GGTTAGTTGGCCCCCTTAAA-3´ | |
| Reverse | 5´-AGTTGAGCGAAAAGGGGATT-3´ | |
| OXA-51 | 353 | |
| Forward | 5´-TAATGCTTTGATCGGCCTTG-3´ | |
| Reverse | 5´-TGGATTGCACTTCATCTTGG-3´ | |
| OXA-58 | 599 | |
| Forward | 5´-AAGTATTGGGGCTTGTGCTG-3´ | |
| Reverse | 5´-CCCCTCTGCGCTCTACATAC-3´ |
3.4. Generation of DNA Fingerprints by RAPD-PCR
PCR reaction mixtures (25 μL) contained 2 μL of template DNA, 0.5 mM each dNTP, 4 mM MgCl2, 1.5 U of Taq polymerase, and 1 μM DAF4 primer (5´-CGGCAGCGCC-3´) in the buffer provided by the manufacturer (CinnaGen, Iran) (16). Amplifications were performed in a thermal cycler (Techne TC-3000G, UK) using the following program: denaturation at 94°C for 2 minutes followed by 45 cycles of denaturation at 94°C for 40 seconds, annealing at 48°C for 1 minute, and extension at 72°C for 40 seconds, with a final extension period of 5 minutes at 72°C. The PCR products were separated on 1.2% agarose gels, stained, and visualized as described above.
3.5. Statistical Analyses
Chi-square analysis was employed for the comparison of OXA gene carriage between the two groups of isolates using the SPSS version 21. Dendrograms for RAPD fingerprints were generated based on the band-matching coefficient of Dice, SAHN, and clustering in the unweighted pair group method with arithmetic averages (NTSYS-pc version 2.02; Exeter Software, Setauket, NY, USA).
4. Results
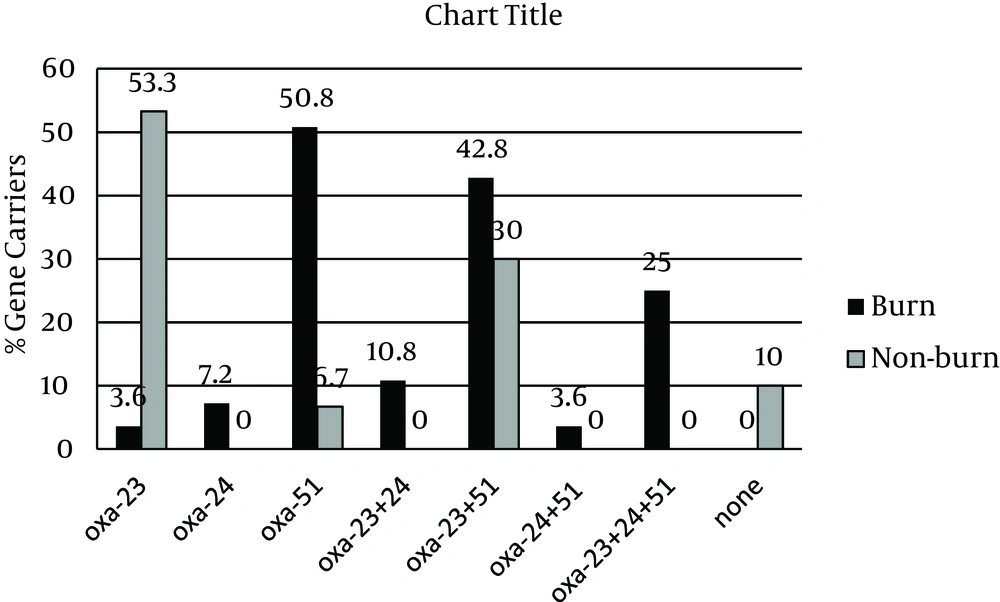
All isolates were resistant to imipenem with MIC and MBC values of > 32 μg/mL. However, these values for non-burn isolates ranged from 32 to 128 μg/mL, whereas the burn isolates had MIC and MBC values of > 128 – 1012 μg/mL, indicating even higher levels of resistance. The results of PCR amplifications (Figure 1) showed that blaOXA-23 was present in 47 isolates (81%), which was not significantly different between the two groups (78.6% burn vs. 83.3% non-burn). The presence of blaOXA-51 was shown in 34 isolates (58.6%), of which most were burn isolates (82.1% vs. 36.6%, P < 0.05). Interestingly, blaOXA-24 was found in 12 isolates (20.7%), all belonging to the burn group, and none of the isolates harbored blaOXA-58 (Figure 1). Overall, among the non-burn group, 16 isolates (53.3%) carried blaOXA-23 and two (6.7%) harbored blaOXA-51 alone, nine (30%) had both genes, and three did not carry any of the tested OXA genes. On the other hand, OXA-like genes were more widely distributed among the burn isolates, where one (3.6%), two (7.2%), and three (10.6%) isolates carried blaOXA-23, blaOXA-24, and blaOXA-51 alone, respectively. Twelve burn strains had both blaOXA-23 and blaOXA-51 (42.8%), two carried blaOXA-23/blaOXA-24 (7.2%), one harbored blaOXA-24/blaOXA-51 (3.6%), and seven isolates had all three OXA-like genes (25%). These results show a higher rate of multiple OXA gene carriage among the burn isolates and correlate with the higher imipenem MIC and MBC values obtained for the burn isolates.
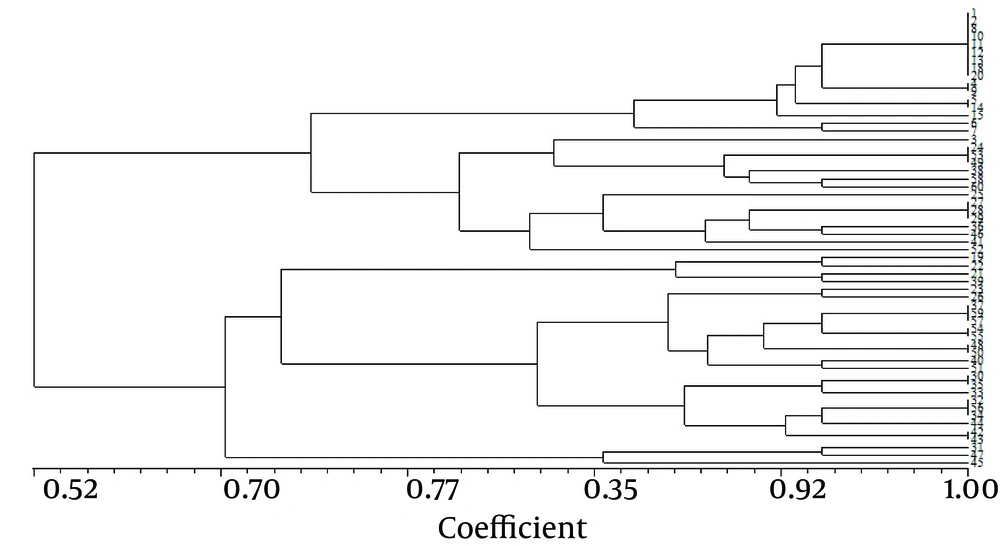
Figure 2 presents the RAPD fingerprints of A. baumannii isolates. Analysis of these fingerprints revealed two clusters (A and B) at a similarity level of 70% and nine groups at a similarity level of 85% (Figure 3). Table 3 shows the distribution of RAPD fingerprints among burn and non-burn isolates. Most of the burn isolates (n = 18, 64.3%) were placed in cluster B, of which, seven, eight, and three belonged to the groups B7, B8, and B9, respectively. Of the 10 burn isolates in cluster A, four, five, and one isolate were placed in the groups A3, A4, and A5, respectively (Table 3). Most of the non-burn strains belonged to cluster A (n = 21, 70%), of which majority (n = 16, 76.2%) were in the group A1 and the remaining five were found in groups A2 (n = 1), A3 and A4 (n = 2 each). The non-burn strains placed in cluster B were found in groups B6 and B7 (n = 4 each), and B8 (n = 1). These results suggest the clonal spread of A. baumannii in the clinical isolates from the two hospitals. However, a higher degree of heterogeneity was observed among the burn isolates compared to the non-burn isolates.
5. Discussion
Nosocomial outbreaks of oxacillinase-producing A. baumannii have been reported worldwide. Among the five main phylogenic subgroups, the chromosomally located blaOXA-51-like gene has been used as a species-specific marker for the identification of A. baumannii (16-18). However, plasmid-mediated OXA-51 has also been detected in A. baumannii isolates (12). In addition, “non-baumannii” species of Acinetobacter have been shown to harbor blaOXA-51 (19). In the present study, 58.6% of the A. baumannii clinical isolates harbored blaOXA-51, majority of which were burn isolates. The prevalence of blaOXA-51 in the clinical isolates of A. baumannii was reported as 77.8% in Turkey and 85.3% in South Africa (20, 21). Iranian studies have mostly used the presence of blaOXA-51 as a tool to identify A. baumannii and have shown 100% carriage of the gene (18, 22, 23). The presence of blaOXA-51 does not correlate with the level of carbapenem resistance because the gene is regulated by the insertion sequence ISAba1; this indicates that resistance to carbapenems cannot be inferred solely from the detection of blaOXA-51-like genes (24). In contrast, genes encoding OXA-23, OXA-24, and OXA-58 enzymes are consistently associated with resistance or reduced susceptibility to imipenem (24). Among oxacillinases, OXA-23 has been shown as the most prevalent gene in Acinetobacter spp. worldwide, followed by blaOXA-51 (11, 25, 26). As found in the present study, several studies from Iran have found blaOXA-23 to be the most prevalent oxacillinase gene, and found much lower rates of blaOXA-24 and blaOXA-58 occurrence among A. baumannii clinical isolates (18, 23, 27).
Coexistence of multiple OXA genes has been reported in many studies. Feizabadi et al. showed that 25% and 17.9% of MDR A. baumannii carried blaOXA-51/blaOXA-23 and blaOXA-51/blaOXA-24, respectively (18). A study from Poland compared OXA gene carriage between burn and ICU isolates and found that blaOXA-51/blaOXA-24 genes were dominant in the ICU group and blaOXA-51/blaOXA-23-like genes were dominant among the burn isolates (28). Our results showed that only 30% of the non-burn isolates carried blaOXA-23/blaOXA-51, while 78.6% of the burn isolates harbored two to three oxacillinase genes.
Our RAPD-PCR results showed clear differences between the distribution of OXA genes in the two groups of isolates; higher heterogeneity was observed among the burn strains (Table 2). Sadeghifard et al. showed 6 RAPD-PCR patterns among Acinetobacter spp. isolates in Tehran and found distinguishable fingerprints for A. baumannii strains compared to the non-A. baumannii isolates (29). Popova et al. used RAPD-PCR to type 130 MDR A. baumannii strains and showed that all strains grouped into two clusters (A and B), of which 82% were in the two dominant groups (30). We also found two clusters among which the RAPD fingerprints of both groups of isolates were distributed. However, most of the burn isolates (64.3%) were placed in one cluster (B). Conversely, 70% of the non-burn isolates were placed in the second cluster (A). In addition, there was much higher heterogeneity among the burn isolates at a similarity level of 85%, compared to that among the non-burn isolates (Table 2). These results show the limited lineage of infection-causing A. baumannii isolates in hospital environments. The heterogeneity, when detected, would most probably be attributed to the horizontal transfer of genetic elements such as plasmids among individual isolates.