1. Background
Staphylococcus aureus is one of the most common causes of infection in both healthy and immune-deficient individuals. The bacterium has various virulence factors, such as staphylococcal super antigens (SAgs) (1). Staphylococcal super antigens are characterized by their ability to make a cross-link between some subsets of T cell receptors and class II major histocompatibility (MHCII) molecules by attaching at different positions of the MHC cleft (2, 3). Different strains of S. aureus can produce different SAgs; however, most strains can produce toxic shock syndrome toxin-1 (TSST-1), staphylococcal enterotoxin B (SEB), and staphylococcal enterotoxin C (SEC) (2). Staphylococcal enterotoxin B is one of the toxins responsible for staphylococcal food poisoning in humans; it acts by stimulating cytokine release and mediates inflammation (4, 5). Owing to its potency and stability under various environmental conditions, Staphylococcal enterotoxin B can cause severe poisoning and pose a threat to human life. Therefore, the detection of this toxin in food and environments is of the utmost importance. Staphylococcal food poisoning is usually diagnosed based on clinical symptoms. Staphylococcal enterotoxin B may be found in the blood, urine, respiratory secretions, and other body fluids.
There are several methods for the identification of staphylococcal enterotoxins, including microbiological methods and testing for toxin production. To optimize the incubation time and the production yield of toxins, various factors, such as pH, osmotic pressure, and the use of substrates, are important (6). Many methods are based on the direct detection of enterotoxins in food, with the ability to detect enterotoxins at the nanogram scale in one gram or milliliter of food (7, 8). Enterotoxins can be detected by enzyme-linked immunosorbent assays, chemiluminescence, or reversed passive latex agglutination tests. Although these conventional methods have acceptable sensitivities, most of them are time consuming; hence, more rapid and sensitive diagnostic methods are required (9-12). Whereas these methods depend on the expression and presence of the toxin in samples, other sensitive and specific methods, such as PCR, can detect enterotoxin-producing bacteria before the production of the toxin. Because the DNA remains intact after heating, PCR-based methods are able to detect genes (13, 14). Sharma et al. (6) reported a multiplex PCR method for the detection of all staphylococcus enterotoxins. They used one universal and five specific primers in a single reaction. This type of one-step PCR is very useful for the detection of different staphylococcal enterotoxin genes. In this study, we performed a comparative analysis to determine the best method for the detection of SEB. Both methods have advantages and disadvantages.
In the present study, the PCR technique was applied to identify the presence of toxin-producing bacteria in samples. The strip assay has been applied for the detection of antibodies (15) and antigens (16, 17) and has been under development for several years. This technique is based on an immunochromatographic procedure that uses Ag-Ab properties and enables the rapid detection of substances. It includes several benefits, such as a user-friendly format, rapid results, and long-term stability over a variety of weather conditions; additionally, in comparison with other methods, it is more economically feasible. Recent studies have used monoclonal antibodies (mAbs) in a strip assay to detect the toxin ricin (17) and a polyclonal antibody (pAb) to detect SEB (18) and botulinum neurotoxin (19, 20).
2. Objectives
In this study, we applied a mAb for the development of a rapid and sensitive sandwich immunochromatographic assay that can detect SEB contamination in food or clinical samples. The aim of the current study was to develop a rapid immunochromatographic test using a lateral flow device (LFD) for the diagnosis of SEB. The PCR technique was applied to confirm the presence SEB gene. Earlier detection of SEB in food will be helpful to screen for food poisoning as well as to decrease gastrointestinal pathological conditions.
3. Materials and Methods
3.1. Bacterial Strains
In this study, 80 samples obtained from dairy products suspected of S. aureus contamination were tested. The samples were collected under the supervision of the Food and Drug Administration of Iran under aseptic conditions and were transported to the laboratory at 4°C. Samples were diluted at a ratio of 1:99 with sterile physiological serum (0.9% NaCl in double-distilled water). For S. aureus isolation, 1 mL of each sample was inoculated into cooked meat medium (Sigma, Germany) containing 9% NaCl and incubated at 37°C for 48 hours (21). Then, 100 µL of culture was spread on Baird-Parker agar, nutrient agar, and mannitol salt agar (MSA) (Sigma, Germany), and incubated at 37°C for 24 - 48 hours. Dark colonies with a clear zone in Baird-Parker agar were selected for further confirmation. The colonies were subjected to catalase, coagulase, DNase, and hemolysis assays.
3.2. Colloidal Gold Conjugation
Based on standard protocols (22), the anti-SEB mAb (Sigma, Germany) was diluted to 10, 20, 50, 100, 250, 500, 750, and 1000 ng/mL in borax buffer (sodium tetraborate, 2 mM) and the pH was adjusted to 9 with 0.2 M K2CO3, then 0.5 mL of colloidal gold (Arista Biological, Allentown, PA, USA) with a pH of 9 was added to 100 µL of each antibody dilution and incubated for 10 minutes at room temperature. Then, 100 µL of 10% NaCl was added to each vial and the color changes were measured. The lowest concentration that resulted in no change in color was considered the optimal concentration for stabilizing the soluble gold particles. In order to stabilize the conjugated antibody, the solution was centrifuged at 8,000 g at 4°C for 30 minutes. Gold-conjugated antibodies were stored in phosphate buffer (20 mM, pH 9), 0.1% Tween-20, and 1% BSA at 4°C.
3.3. Production of the Immunochromatographic Strip for Rapid Testing
Conjugated mAb was diluted in 20 mM phosphate buffer and applied to the conjugate pad in 2 µg/cm using an Isoflow Dispenser (BioDot, Irvine, CA, USA). Two test strips were developed. The test line contained 1 µg/cm SEB (Sigma) for the competitive method and the anti-SEB antibody (HyTest, Turku, Finland) at a concentration of 0.5 µg/cm for the sandwich method. For both methods, the control line contained 0.5 µg/cm goat anti mouse antibody (HyTest). Strips contained various layers, including the sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad (Schleicher and Schuell, Dassel, Germany), and were assembled on a backing card (G & L, San Jose, CA, USA) with a 2-mm overlap on each sequential layer. As shown in Figure 1, all strip sheets were cut to a width of 5 mm with an automated cutter.
3.4. Extraction of Genomic DNA
DNA was extracted from Staphylococcus aureus type S-6 (NCTC) using the Pure Link Genomic DNA Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
3.5. Confirmation of the Strains Using PCR
Confirmed S. aureus colonies were used to extract genomic DNA. The sequence of the SEB gene was obtained from GenBank (AY852244.1) and primers were designed by GeneRunner to specifically identify the presence of the SEB gene in samples. These primers were predicted to result in the amplification of a 633-bp DNA fragment and are as follows:
Forward primer, 5’-ACACCCAACGTTTTAGCAGAGAGTCA-3’;
Reverse primer, 5’-TCCTGGTGCAGGCATCATGTCA-3’.
PCR was carried out in a 25-µL reaction according to the methods described by Williams et al. (23), with some modifications. The reaction contained the following: 2.5 µL of PCR buffer (10X), 2.5 μL of dNTPs (2 mM), 1.25 µL of forward primer (10 pmol), 1.25 µL of reverse primer (10 pmol), 0.5 µL of Taq DNA polymerase (5 U/μL), 1.5 μL of Mg2+ (50 mM) (CinnaGen, Tehran, Iran) and 50 ng of template DNA. The final volume was adjusted to 25 μL by adding sterile ultrapure water. The thermo-cycling conditions were as follows: 5 minutes at 95°C and 35 cycles of 45 seconds at 95°C, 45 seconds at 60°C, and 45 seconds at 72°C, followed by 10 minutes at 72°C.
3.6. Identification of the SEB Toxin Using the Strip Assay
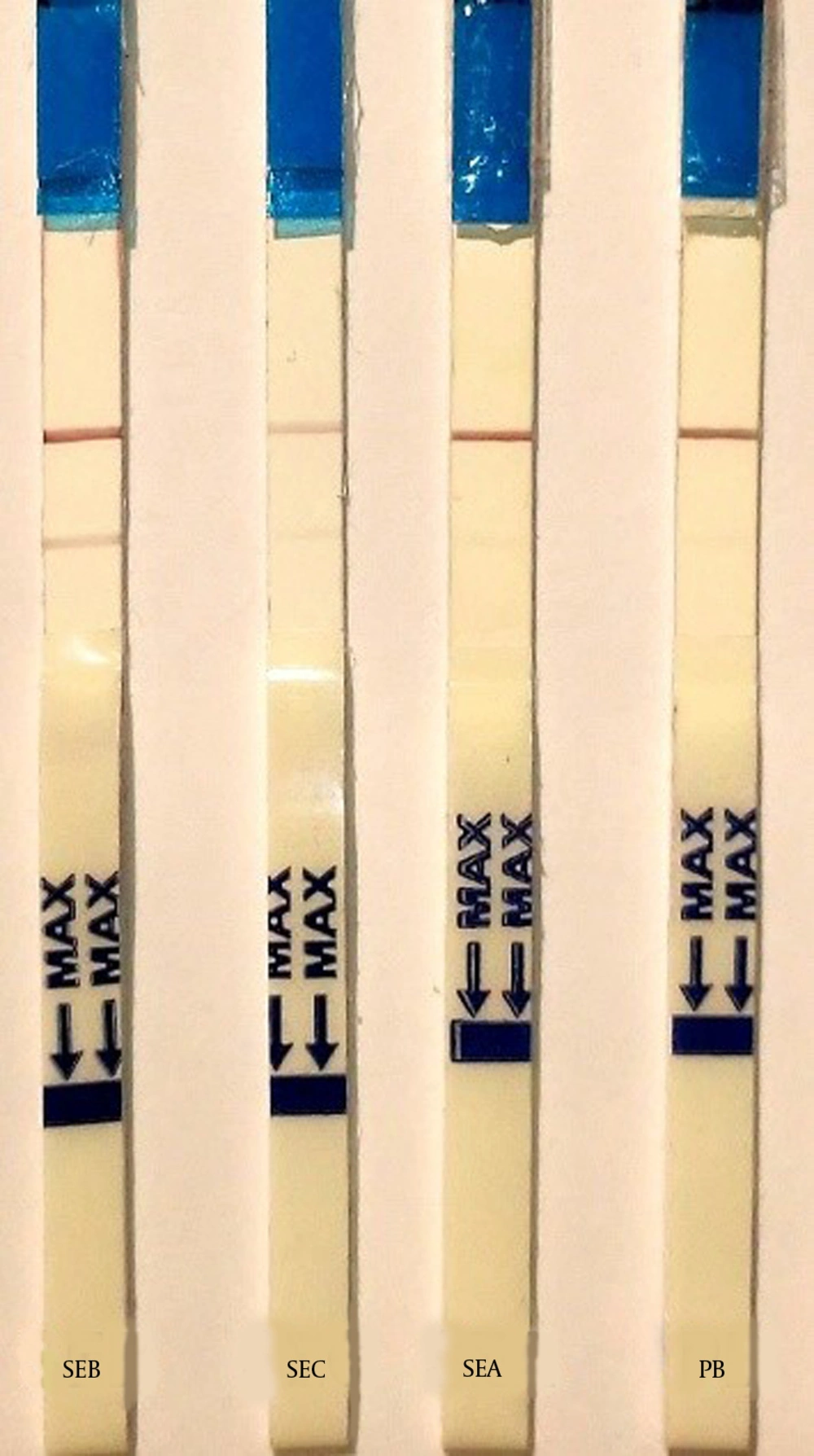
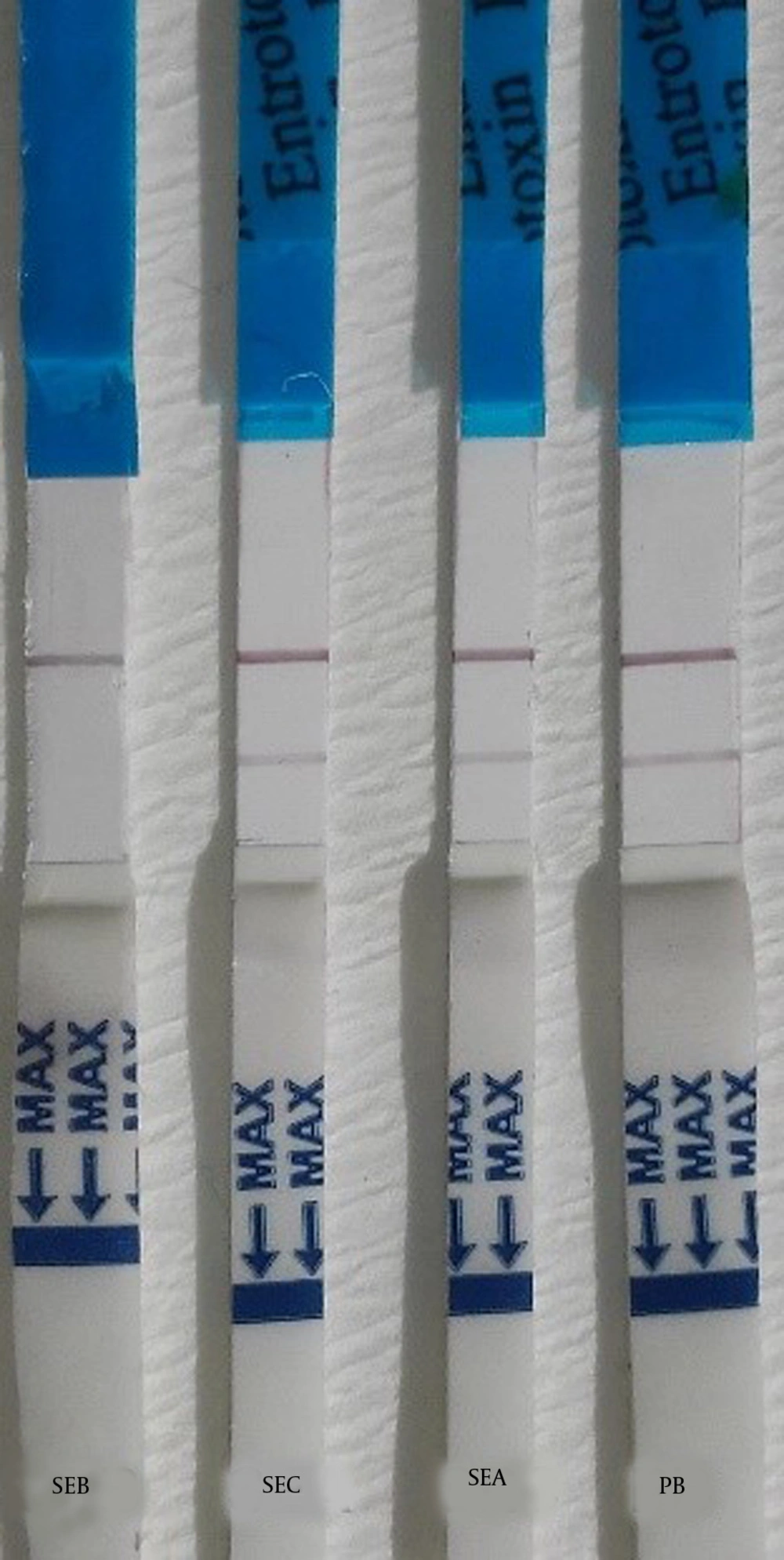
Isolated samples were tested for the presence of the SEB toxin using the strip test assay. As a positive control, 1 µg/mL SEB was used, and as negative control, phosphate buffer was used. Additionally, in order to estimate the sensitivity of the strip tests, serial dilutions of the SEB toxin were prepared, and strips were assayed using different samples, such as SEA, SEB, and SEC samples.
4. Results
In this investigation, 80 samples were isolated from dairy products; samples were processed to confirm the presence of S. aureus. They were cultured in cooked meat medium and on Baird Parker agar, nutrient agar, and MSA. Based on the appearance of a yellow color in the MSA medium and dark colonies with a clear zone in Baird Parker agar, and based on the results of catalase, coagulase, DNase, and hemolysis assays, 54 samples of S. aureus were confirmed.
4.1. Detection of SEB by the Sandwich Method
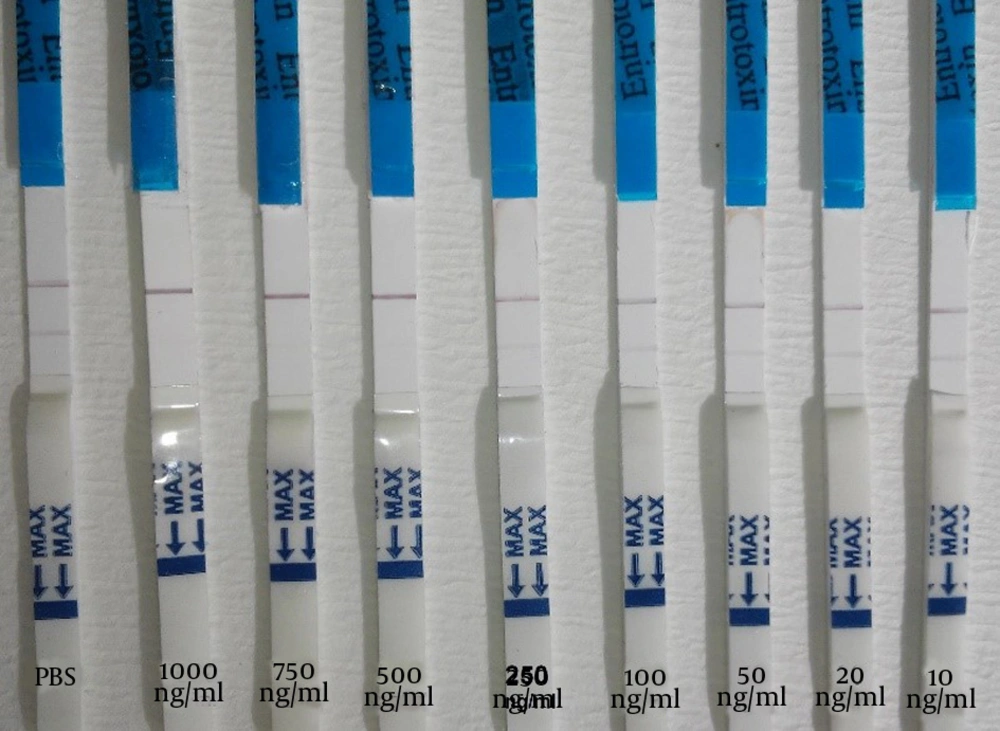
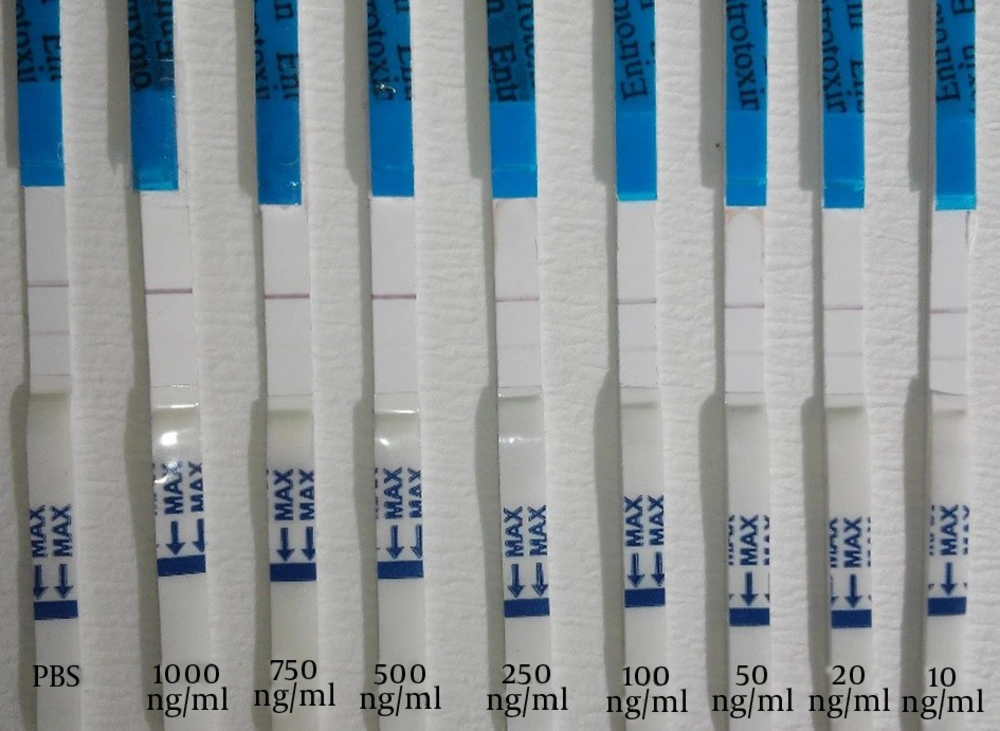
In order to detect SEB, the mAb was captured in a test line (0.5 µg/cm). Conjugated mAb was applied to the conjugate pad at a concentration of 2 µg/cm using the Isoflow Dispenser. The control line was coated with 0.52 µg/cm goat anti mouse antibody. The strips were used to detect SEB at various concentrations; the intensity of the red color in the test line was proportional to the SEB concentration and the control line should visualized at any time. The sensitivity (Figure 1) and specificity (Figure 2) of the sandwich method were 91% and 84%, respectively (Table 1). The detection limit of the SEB strip for the sandwich method was 10 ng/mL.
Detection of SEB by the competitive method for the competitive method, the test line contained 1 µg/cm SEB the mAb-conjugated (2 µg/cm) and the goat anti mouse antibody, as control line, were applied to the conjugate pad and was visualized by a red line. As previous described, and nitrocellulose membrane, respectively. In the positive control (SEB 10 µg/mL), only the control line resulted in a visible red line. In the negative control (PB), two red lines were observed on the test strip. The sensitivity (Figure 3) and specificity (Figure 4) for the competitive method were 77% and 68%, respectively (Table 1). Additionally, the detection limit of the SEB strip for the competitive method was 250 ng/mL.
4.2. Comparison Between the Strip Assay and PCR Method
Two strip assays were designed, one based on the sandwich method and another based on the competitive method. Based on the two strip assays, the sandwich assay had a higher sensitivity. Accordingly, it was chosen for additional analyses and the results were compared with those of the PCR-based method. The sandwich strip assay detected SEB contamination in 8 of 23 total samples. The PCR method detected 9 samples contaminated with SEB from the same pool of samples. Eight of those were positively identified by the sandwich strip assay. These results suggest that the strip assay is able to detect contamination with almost the same accuracy as that of the PCR method; it is much faster and does not require upstream processing.
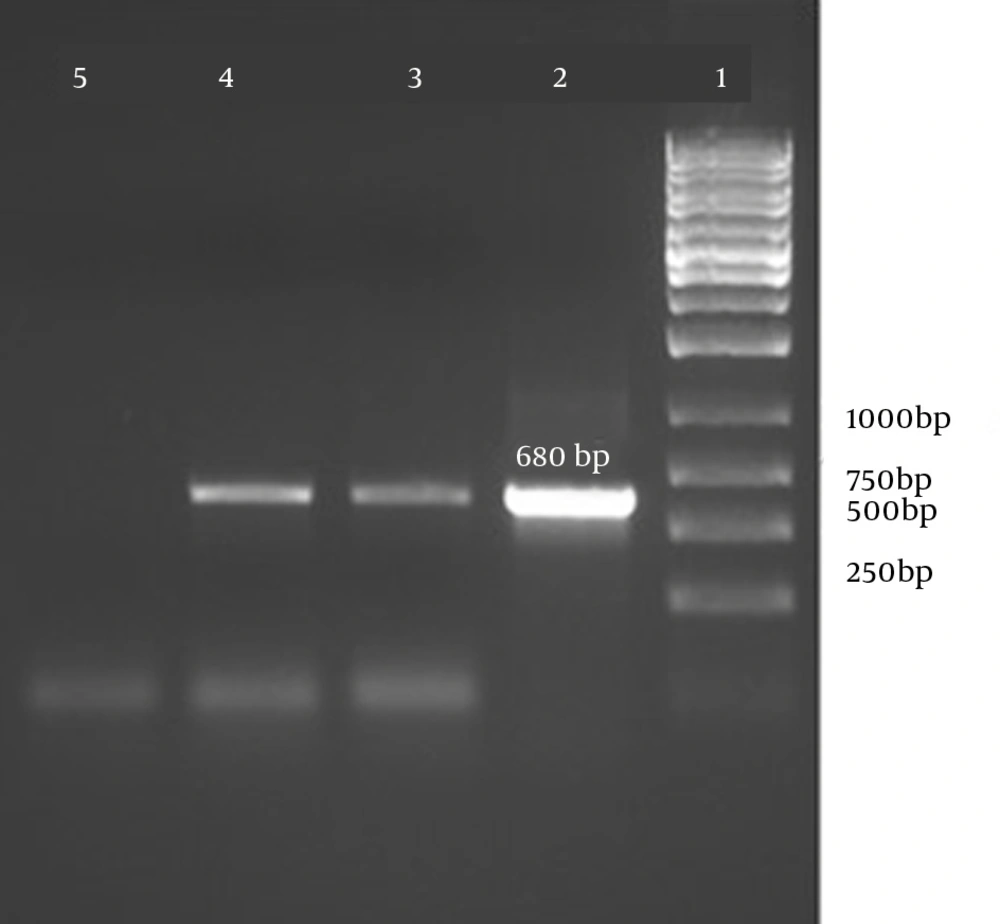
Detection of SEB by PCR The presence of the SEB gene was confirmed by PCR and the amplicons were run on a 1% agarose gel. The results are shown in Figure 5.
5. Discussion
In the present study, we used the colloidal gold-labeled antibody, which is among the most broadly employed strategies for constructing an LFD, to detect SEB. SEB is an important member of the staphylococcal enterotoxin group, which causes clinical complications and food poisoning in humans. Given its potency and ability to cause illness at very low concentrations, various techniques have been developed to improve the detection of this toxin. Sensitivity, specificity, and assay time are among the most important factors that are considered in the design of such assays (24, 25). The use of gold-conjugated antibodies is economically feasible, and they have high stability in dry conditions (24 months in the present study) (22). Furthermore, in a comparison with other conjugation strategies, the colloidal gold-Ab conjugation is easier and results in less denaturation and deactivation of the antibody (26, 27).
Two strip assays were designed, one based on the sandwich method and another based on a competitive method. The sandwich assay had a higher sensitivity and was chosen for a comparative analysis with the PCR-based method. These results suggest that the strip assay is able to detect contamination with almost the same accuracy as that of the PCR method in much less time and without the need for upstream processing. The PCR technique is a prevalent method used to confirm the presence of the SEB gene and was compared with the LFD method. In total, 54 samples that were verified by PCR were considered positive and 26 remaining samples were negative using the immunoassay strip tests. For the negative samples, the results obtained using the PCR and immunoassay tests were similar, but for the positive samples, there were some differences related to differences in the diagnostic principles.
The higher sensitivity of the sandwich method was attributed to the use of two specific anti-SEB antibodies, whereas the competitive method used only a single anti-SEB antibody; the sensitivities were 10 ng/mL and 250 ng/mL, respectively. Over the past decade, LFDs have been the most well-known commercial rapid immunochromatographic tests. They are simple, inexpensive, fast (fewer than 5 minutes for diagnosis), do not require expert personnel, and require a small sample volume (200 µL); these advantages make LFD ideal for initial screening tests (22, 28-30). There is another immune-based method for the detection of SEB toxins that uses labeled anti-SEB polyclonal antibodies. The sensitivity of this technique depends on the time of incubation and the results are often inconsistent. However, the strip assay is fast and depends on the travel time of the Ag-Ab complex through various layers of the strip. The use of more sensitive and specific antibodies in these devices will increase the sensitivity of the assay, and minimal concentrations of toxins can be detected in a much faster period of time. Another method that is widely used for the detection of SEB toxins is the PCR-based technique. This method is based on the detection of the toxin-producing gene in pathogenic strains of bacteria. PCR is a sensitive method, but requires upstream processes, such as microbiologic methods and genome extraction.