1. Background
Pseudomonas aeruginosa is a Gram-negative rod and belongs to the glucose non-fermentative bacteria family. It is an important opportunistic pathogen that causes infections in patients with weakened immune system and cystic fibrosis genetic defect, as well as patients at Intensive Care and Burn Units (1). These bacteria have a high resistance to a wide range of existing antimicrobials and antibiotics. Eradication of infections by P. aeruginosa is difficult due to their inherent resistance to a wide range of antibiotics, as well as their mobile genetic elements including resistance genes (2).
Emergence of carbapenem-resistant strains has expanded amongst Gram-negative bacteria in the recent years. The most important mechanisms of resistance to carbapenems are production of carbapenemase enzymes and secretory pumps, and lack of expression of external membrane proteins (porins) (3). Carbapenemases have the ability to hydrolyze penicillins, cephalosporins, monobactams and carbapenems (4). The presence of KPC in transferable plasmids and transposons can result in a rapid spread among bacteria including Klebsiella pneumoniae, Enterobacter species, Acinetobacter baumannii and P. aeruginosa (5). The world’s first report of KPC-2 presence in Pseudomonas was reported by the International Center of Medical Research and Education in Columbia, during year 2006. Subsequently, a study from Puerto Rico reported on two types of KPC (KPC-2 and KPC-5) in P. aeruginosa (6).
Rapid detection of strains producing carbanapemases, such as KPC, is crucial for preventing incorrect treatment, controlling hospital infections, and preventing the spread of these strains. Modified Hodge Test (MHT) proposed by the Clinical and Laboratory Standards Institute (CLSI), and boronic acid inhibitor-based assay are among the many phenotypic methods used in microbiology laboratories for identification of Group A carbanapemases in the Enterobacteriaceae family (7).
2. Objectives
In Iran, no study has so far been carried out for evaluation of KPC frequency using phenotypic tests of MHT and boronic acid inhibitor-based assay and for their comparison with the genotypic method of Polymerase Chain Reaction (PCR) in P. aeruginosa. Therefore, the present study was performed to evaluate the frequency of KPC in P. aeruginosa, isolated from clinical samples of educational hospitals of Arak University of Medical Sciences, using the mentioned phenotypic and genotypic methods.
3. Materials and Methods
This project was approved by the Arak University of Medical Sciences Ethical Committee (No. 92-152-14).
3.1. Bacterial Isolates and Detection of Extended Spectrum β-Lactamases
This study was a descriptive cross-sectional study carried out from August 2012 to July 2013, in which 108 P. aeruginosa isolates were collected from urine, sputum, and ulcer samples of hospitalized patients at the educational hospitals of Arak University of Medical Sciences, Arak, Iran. Pseudomonas aeruginosa was identified using standard biochemical methods, including Gram staining, catalase test, Oxidative-Fermentation (OF) test, triple sugar iron agar (Merck, Germany), growth at 42°C, and production of blue-green pigments. Antibiotic susceptibility pattern of isolates was also determined using the disk diffusion method (Kirby-Bauer), according to the standards of Clinical and Laboratory Standards Institute (CLSI) (8). All disks were made by the MAST Company (UK) and included the following, ceftazidime, ciprofloxacin, amikacin, gentamicin, imipenem and meropenem. The standard strain of P. aeruginosa ATCC 27853 was used as the antibiogram control. An intermediate growth inhibition zone was considered as resistant.
3.2. Modified Hodge Phenotypic Test
First, a suspension of the standard strain of Escherichia coli ATCC 25922 was prepared at a concentration of 0.5 McFarland and then diluted to 0.01 McFarland with normal saline. The suspension was then lawn cultured on Mueller Hinton medium (Merck, Germany) using a swab. A 10-μg meropenem disc (Mast,UK) was then placed on the centre of the plate. Pseudomonas aeruginosa sample was cultured in a direct line from the disc edge towards the plate edge and the plates were incubated at 37°C for 16 to 24 hours. Cloverleaf-like structure indicated the production of carbapenemase (9).
Since KPC-producing P. aeroginosa inhibit E. coli on Muller Hinton medium and lead to unexplained results, E. coli standard strain and K. pneumoniae ATCC 700603 were cultured on separate Mueller Hinton medium and Pseudomonas was cultured on and the results of the two tests were finally compared (10).
All isolates were tested for KPC carbapenemases production on disks containing boronic acid. A disk containing imipenem (Mast,UK) and another containing imipenem with 400 μg of boronic acid (Sigma-Aldrich, Germany) were placed on the agar. The diameter of the growth-inhibitory zone around the imipenem disk with boronic acid was compared with that around the corresponding imipenem disk without boronic acid. The test was considered positive for the detection of class A carbapenemase production when the diameter of the growth-inhibitory zone around the imipenem disk with boronic acid was ≥ 5 mm larger than that around the disk containing the imipenem substrate alone (11).
3.3. Polymerase Chain Reaction (PCR)
DNA was extracted from bacterial samples using the boiling method. The blaKPC gene was detected by PCR using the following specific primers; Forward: 5'CGTCTAGTTCTGCTGTCTTG3' and Reveres: 5'CTTGTCATCCTTGTTAGGCG3' (12). The DNA amplification, programmed for the blaKPC gene, consisted of an initial denaturation step (94˚C, three minutes), followed by 40 cycles of denaturation (94˚C, 30 seconds), annealing (58.2˚C, 60 seconds) and extension (72˚C, 30 seconds), and a single final extension (72˚C, five minutes).
4. Results
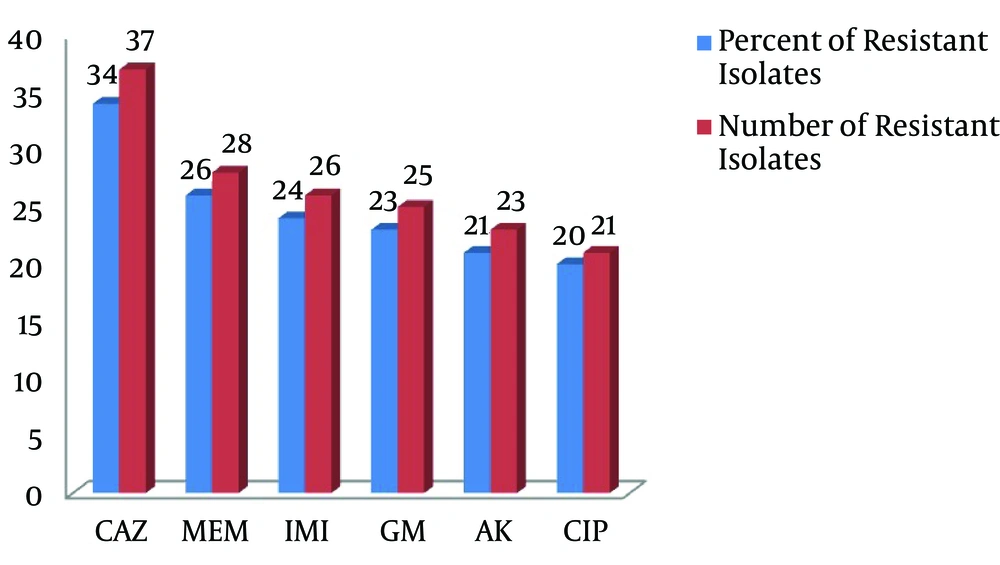
Overall, 108 isolates of P. aeruginosa were collected from hospitalized patients’ clinical samples at three educational hospitals of Arak University of Medical Sciences. The results of the antibiotic sensitivity pattern (number and percentage of resistant isolates) of P. aeruginosa isolates, obtained through the disk diffusion method, are presented in Figure 1.
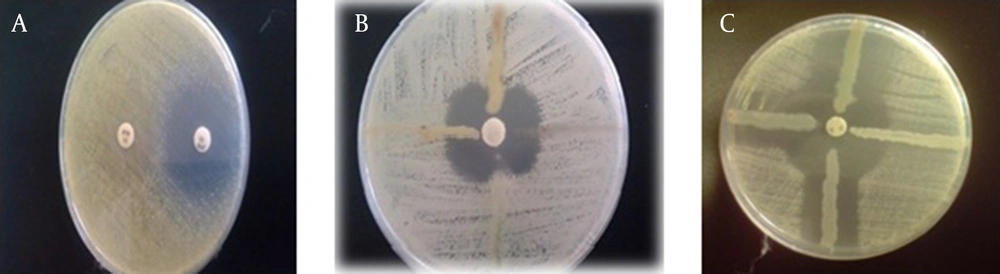
Assessment of carbapenemase production in P. aeruginosa through the MHT method, using the standard strain of E. coli ATCC 25922, resulted in only 27 examinable isolates; 81 isolates were unexplainable due to Pseudomonas inhibition of E. coli growth in the culture medium (Figure 2A). However, out of 108 isolates examined by the MHT test, using the standard strain of K. pneumoniae ATCC 700603, 38 isolates of P. aeruginosa produced carbapenemase and the results of this test were negative for 70 isolates (Figure 2B).
A, modified hodge test (MHT). Results for three Pseudomonas isolates, which were unexplainable when using E. coli ATCC 25922 standard strain. B, MHT test results using K. pneumoniae ATCC 700603 standard strain. C, The results of combination-disk test with boronic acid, which was positive for the examined strains.
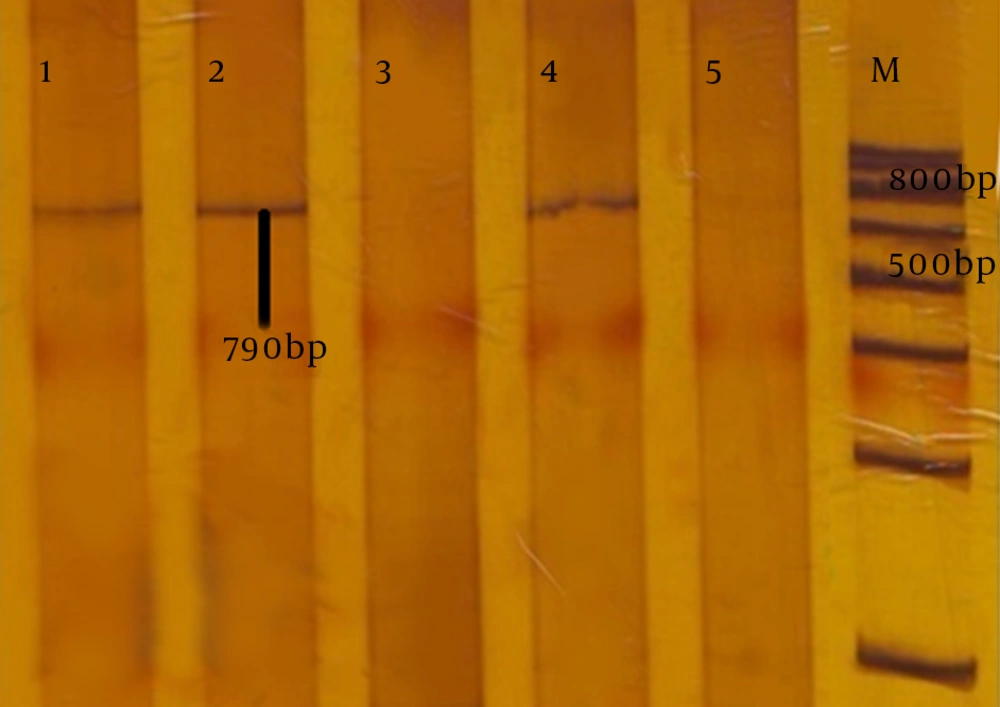
Using the combination-disk test with boronic acid, 26 isolates were identified as carbapenemase-producing strains, while 82 isolates were negative (Figure 2C). The characteristics of 13 isolates are presented in Table 1. Of the 108 isolates of P. aeruginosa studied by PCR, 13 isolates (12%) had the KPC carbapenemase encoding gene (blaKPC) (Figure 3).
| Number of Isolate | IMI | MEM | PCR for KPC | MHT | Boronic Acid |
|---|---|---|---|---|---|
| 1 | S | S | + | + | + |
| 2 | R | R | + | + | + |
| 3 | S | S | + | + | + |
| 4 | R | R | + | + | + |
| 5 | R | S | + | + | + |
| 6 | S | S | + | + | + |
| 7 | R | S | + | _ | _ |
| 8 | S | S | + | + | + |
| 9 | R | R | + | _ | + |
| 10 | S | R | + | + | _ |
| 11 | S | S | + | _ | + |
| 12 | S | S | + | + | + |
| 13 | R | R | + | + | + |
a Molecular studies detected 50% blaVIM-1, 56.6% blaVIM-2, and 6.6% blaIMP-1 while none of the strains had blaIMP-2 and blaSPM-1 genes.
Lane M is the marker (DNA ladder, 100bp); lanes 1, 2 and 4, are amplified products of KPC gene in P. aeruginosa, isolated from hospital samples. Lane 5, is the amplified product of the negative control K. pneumoniae lacking the KPC gene. Lane 3, the amplified product of the KPC gene in P. aeruginosa lacking the KPC gene.
5. Discussion
Utilization of reliable methods for identifying carbapenemase-producing strains and determining their antibiotic resistance pattern could have a very important role in treatment of infections caused by these strains. This could be an important step in the control of hospital infections, in order to prevent patients’ mortality and to reduce health care costs (3, 13). Since no study has so far been carried out for the evaluation of the frequency of P. aeruginosa-producing KPC carbapenemase in Iran, in the present study, the presence of KPC carbapenemase was examined in P. aeruginosa isolated from clinical samples of patients hospitalized at hospitals of Arak University of Medical Sciences, using the genotypic method of PCR. The efficiency of MHT and boronic acid phenotypic methods for identification of KPC enzyme was compared with the PCR.
Due to their wide range of activities, carbapenems are commonly used to treat infections caused by multidrug-resistant bacteria, particularly Extended-Spectrum Beta-Lactamases (ESBLs)-producing bacteria and AmpC enzyme-producing strains (14). Spread of carbapenemase-producing bacteria throughout the world in the recent years has been considered as a major threat to public health. Among carbapenemases, KPC has a high frequency and has been commonly found in K. pneumoniae and Enterobacter (15). The enzyme is encoded by a gene located on mobile elements such as plasmids; this results in KPC gene transfer among bacteria. Furthermore, the KPC enzyme hydrolyses beta-lactam drugs, including monobactams, carbapenems, and third-generation cephalosporins (16). The presence of this enzyme in P. aeruginosa was first reported in Colombia, during year 2007 (17) and subsequently in other countries such as Puerto Rico (18), Trinidad and Tobago (19), the United States, and China (3, 6). Recent studies from Iran have also indicated the prevalence of KPC-producing bacteria (20). The highest prevalence of KPC gene in Iran, examined by PCR, was reported in K. pneumoniae by Hashemizadeh et al. as 11.9% (21). In the present study, 13 isolates (12%) of P. aeruginosa were positive for KPC, using PCR. This could indicate horizontal transfer of KPC genes among bacteria isolated from nosocomial infections in Iran. The prevalence of KPC in P. aeruginosa was 4.1% in Puerto Rico; this was lower than the present study (22). The pattern of antibiotic resistance in 13 KPC-producing isolates in the present study was such that they were all resistant to ceftazidime, gentamicin, amikacin, and ciprofloxacin. Thus, carbapenemase detection cannot be performed by antibiotic profiles in the laboratory. Phenotypic resistance caused by carbapenemase is affected by many factors such as strain, expression level, type, or enzyme variant, as well as the presence of other resistance mechanisms such as reduced permeability, secretory pumps, and other activities of beta-lactamases (4). In the present study, the disk diffusion method showed that of 13 KPC-producing isolates, six and five were resistant to imipenem and meropenem, respectively.
The Modified Hodge Test is a sensitive phenotypic method for detection of carbapenemases, however, it cannot identify the enzyme type. Instead of the standard strain of E. coli ATCC 25922, Pasteran et al. (10) used the standard strain of K. pneumoniae ATCC 700603 (lacking carbapenemases and sensitive to carbapenem) for the MHT. When the common indicator strain (E. coli ATCC 25922) was used, P. aeruginosa inhibited the growth of E. coli standard strains; this resulted in an unexplainable test and decreasing of the test sensitivity and specificity. This problem was resolved through exchanging the indicator strain with the standard strain of K. pneumoniae ATCC 700603. Of the 13 KPC-positive isolates obtained using the MHT method, 10 isolates were considered as KPC-producing strains. The results of the phenotypic method of boronic acid inhibitory effect on Group A carbapenemases identified 11 isolates, out of 13 KPC-positive isolates. The important point in interpretation of the results of phenotypic methods applied in the present study was their positivity in the strains being KPC negative in PCR. This represents the possible presence of other carbapenemase groups such as GES enzymes and various groups of metallo-beta-lactamase in the mentioned strains (given their resistance to imipenem and meropenem).
As a result, unlike the study carried out by Azimi et al. in Iran (20), who only used the MHT phenotypic method to assess the prevalence of KPC-producing strains, using only the MHT method and its positivity in the examined isolates cannot be a reason for the presence KPC, and positive results can be caused by other carbapenemases, and in some cases, by CTX-M ESBLs along with a decrease in porins expression. Therefore, a definite confirmation of KPC-producing strains requires PCR followed by sequencing. The boronic acid method can detect KPC-producing strains, as well as GES amongst isolates. In isolates with AmpC overexpression and change in porins, boronic acid method may lead to false-positive results (9, 23). Comparison of the two phenotypic methods used in this study showed that boronic acid is more sensitive than MHT in identification of KPC-producing strains (84.6% vs. 77%).
Given the significant prevalence of KPC-producing P. aeruginosa and their drug multi-resistance, proper measures seem necessary for controlling infections of these strains, such as their rapid and reliable detection and administration of appropriate antibiotics for treatment. In future studies, the origin of infection and genetic relationships between strains obtained from different hospitals can be evaluated through epidemiologic molecular approaches. A substantial amount of P. aeruginosa isolated from clinical samples of hospitals of Arak (Iran) produced KPC carbapenemase. Due to their low specificity, MHT and boronic acid phenotypic methods cannot completely indicate KPC-producing P. aeruginosa. The sensitivity of boronic acid phenotypic method in detection of KPC was higher than MHT (84.6% vs. 77%).