1. Background
Interactions among organisms are central to understanding any ecosystem. Soil environment is not an exception, however biotic interactions dominating soil biology differ from those in other systems because of the dominating role of some organisms and the lack of autotrophy in soil (1).
Bacillus thuringiensis are Gram-positive soil bacteria and the most successful biological control agent; however, are highly sensitive organisms to various environmental factors including germicidal effect exerted by sunlight, which has been considered as the main factor in the low persistence of vegetative cells and spores of B. thuringiensis in the environment.
Nevertheless, previous studies have shown that the survival of B. thuringiensis IPS82 is also affected by its interaction with other native soil microorganisms in vitro, particularly, by cohabitation with Pseudomonas fluorescens (2). The mechanisms by which P. fluorescens inhibits B. thuringiensis are unknown, yet the most described as the main mechanism of pathogen inhibition is by the production of secondary metabolites (3, 4) and siderophores, which may also play a role in the microbial antagonism (5, 6). This antagonism can be increased in response to several changes of conditions such as the addition of nutrients to the culture medium and the temperature change (7). Therefore, B. thuringiensis IPS82 has little ability to survive as a vegetative cell in interaction with other microorganisms.
2. Objectives
In this work, the population dynamics of B. thuringiensis and its interaction with P. fluorescens in soil was analyzed under different conditions. To follow the populations of both bacteria in soil, microbiological and molecular methods were used, the latter is a useful tool for the detection and monitoring of bacteria and microorganisms that cannot be cultured (8).
3. Materials and Methods
3.1. Bacterial Strains
The B. thuringiensis and P. fluorescens strains used in this study were B. thuringiensis IPS82 (Bacillus Genetic Stock Center), B. thuringiensis recombinant strain SR08, which has a fragment of the plasmid pES8 inserted in its chromosome (9), and Pseudomonas fluorescens ATCC 49838 (Microbiologics Labs. St. Cloud MN USA).
3.2. Culture Media Used
In the present investigation we used the Tris G and Luria Bertani (LB) media (2) and the chemical salt employed in the media were from Sigma-Aldrich.
3.3. Evaluation of the Effect of Nutrients Addition and Moisture Adjustment on Bacterial Interaction Between Bacillus thuringiensis IPS82 and Pseudomonas fluorescens in Soil
Laboratory conditions were designed to achieve a model of bacterial interaction at at a controlled environment and to depict a series of natural conditions of survivors and antagonists between pathogen related bacteria. Primarily, 276 g of soil (with low content of organic material) held in a plastic vase, was sterilized by wet and dried heat to reduce microbial population and distilled water was added to maintain the moisture of the soil. Soil conditions were adjusted to a humidity of 80% or 40%, followed by the addition of 0.05% glucose and 0.05% yeast extract. The proportions of inoculated bacteria in the soil were: B. thuringiensis IPS82 and P. fluorescens: A (105: 10 colony-forming units [cfu]), B (103: 10 cfu), C (105: 105 cfu), D (10: 103 cfu) and E (10: 105 cfu). Control proportions for B. thuringiensis IPS82 were F (103 cfu) and G (105 cfu), and for P. fluorescens were H (103 cfu) and I (105 cfu). Nine proportions were used, with four replicates; therefore 36 vases were used for each treatment, giving a total of 144 vases. All of these vases were inoculated with the above-described proportions of microorganisms and were held at 25°C. The time period for the evaluation was 90 days. Each week 1 g of soil from the four replicated vials was blended with 3 mL of distilled water and diluted suspensions, surface-plated on LB plates with and without the chloramphenicol antibiotic (5 μgmL-1). Plates were incubated at 28°C for 12 hours and colony-forming units (cfu) were counted for B. thuringiensis IPS82 or P. fluorescens.
3.4. Analysis of Bacillus thuringiensis Sporulation Percentage in Soil at Ten and Ninety Days
The percentage of sporulation of the inoculated B. thuringiensis proportions in soil was measured on the ninetieth day of experiment in soil. The rate of sporulation of B. thuringiensis IPS82 and SR08 was also measured on the tenth day. To determine the number of B. thuringiensis vegetative cells or spores, inoculated in soil at each of the different proportions, samples of 1 g from soil were blended in 3 mL of Tris G medium. Next, an aliquot of 100 µL was taken to determine the amount of vegetative cells in the sample, and from this aliquot serial dilutions were made (10-1 to 10-6). All dilutions were inoculated on solid Tris G medium in Petri dishes, incubated at 28°C for eight hours, and counted as colony-forming units (cfu). Only for the selection of spores with respect to the remaining vegetative cells, the tubes with blended soil were incubated at 65°C for 45 minutes, serial dilutions were made and inoculated in solid Tris G medium. The incubation conditions were the same for vegetative cells. Finally the counting of colony forming units was performed.
3.5. DNA Direct Extraction From Soil Inoculated With Bacillus thuringiensis IPS82 and Pseudomonas fluorescens
Samples of 1.5 grams of soil were blended with 6 mL of Tris G medium and incubated in the horizontal position for six hours at 30°C. They were then centrifuged at 3000 rpm for five minutes, and 50% of the volume, free of soil, was separated and the rest with the soil was discarded. To the separated supernatant, 0.6 g of Polyvinylpolypyrrolidone (PVPP), one volume of lysis buffer (50 mM Tris, 50 mM Ethylene diamine tetra acetic acid (EDTA), 1 M NaCl pH 8) and 100 μg of lysozyme were added and the resulting solution was incubated for 30 minutes at 37°C. Subsequently, 20% Sodium Dodecyl Sulfate (SDS) was added (1 mL per 10 mL volume), samples were incubated at 65°C for one hour and shaken by inversion every 15 minutes, centrifuged at 3000 rpm for five minutes, and to 500 μL of the supernatant, 0.1 volume of 3 M sodium acetate pH 7 and 1 volume of isopropanol were added. The samples were incubated at -20°C for 10 minutes and were centrifuged at 13000 rpm for 10 minutes. Finally, the pellet of DNA was resuspended in 100 μL of TE (10 mM Tris 1 mM EDTA). The chemical compounds employed were from sigma-Aldrich (USA).
3.6. Detection of Populations of Bacillus thuringiensis ISP82 and Pseudomonas fluorescens Inoculated in Soil by Molecular Methods
For the detection of populations of B. thuringiensis ISP82 and P. fluorescens in soil, molecular methods such as Polymerase Chain Reaction (PCR) reaction and dot-blot Hybridization were used. To carry out the PCR reaction, 120 μg of DNA, extracted directly from soil as described above, was used. To specifically detect B. thuringiensis ISP82, the cry10Aa gene of B. thuringiensis was used as a probe. These primers were designed in the laboratory, based on the sequences of three conserved motifs encoding the Cry proteins of B. thuringiensis, and the oligonucleotide sequences were: Cry10Aa (Accession number M12662) 5’TTTTGCTGCCCCTGTCTTAG3’ and 3’TAGTGGGTTTAGGTGCGAGA5’ (IDT Technologies, México). The amplification conditions were: denaturation at 94°C for five seconds, annealing at 50°C for 30 seconds, and extension at 65°C for one minute (30 cycles). The PCR product with expected size of 1290 bp was run on agarose (Invitrogen, USA) gel at 0.8%. To carry out dot-blot hybridization, 100 or 120 ng of DNA, extracted directly from soil, was denatured at 95°C for ten minutes and placed as drops on a nylon membrane (Merck Millipore, USA). These membranes were hybridized with a probe of total genomic DNA of B. thuringiensis IPS82 or P. fluorescens marked with 32P (Merck Millipore, USA) and the autoradiography was obtained using Kodak® X-ray films, according to Sambrook et al. 1989 (10).
4. Results
4.1. Evaluation of the Effect of Nutrients Addition and Moisture Adjustment on Bacterial Interaction Between Bacillus thuringiensis IPS82 and Pseudomonas fluorescens in Soil
With the purpose of evaluating the population dynamics of B. thuringiensis and its interaction with P. fluorescens, under different conditions in soil, the effect of the nutrients addition and moisture adjustment was analyzed with the proportions of inoculated bacteria described in the materials and methods. These proportions were chosen based on the previous laboratory works, which were designed to analyze the effect of the interaction between B. thuringiensis and P. fluorescensin vitro. The results determined that there was an antagonistic effect when these bacteria interacted, thus it was interesting in the present work to analyze whether such event was also present in soil.
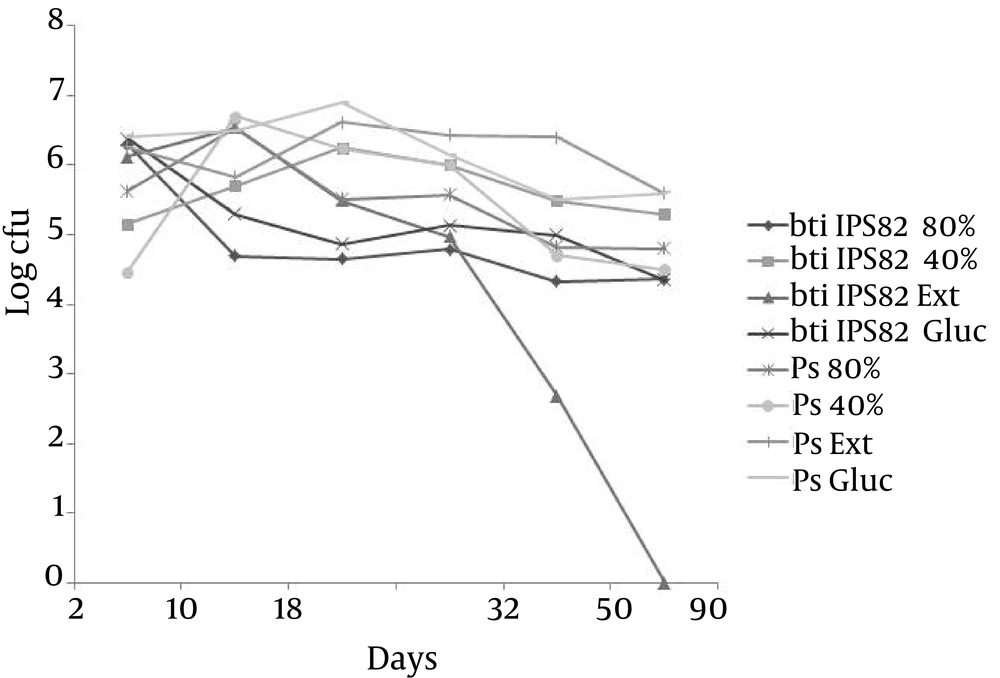
The results of the soil analysis showed that when a high proportion of the population of B. thuringiensis IPS82 with respect to P. fluorescens in A proportion (105: 10 cfu), only in the treatment with the addition of yeast extract, was inoculated, there was a disappearance of the population of B. thuringiensis on the sixtieth day, while in the rest of the treatments, B. thuringiensis IPS82 was recovered even on the ninetieth day. For P. fluorescens in the same proportion, two days after being inoculated, the bacterial population increased four or five orders of magnitude in different treatments (Figure 1). Thus, moisture adjustment had no effect on both bacterial populations while the addition of yeast extract had an effect on the B. thuringiensis population because it disappeared by the sixtieth day. In the B proportion (103: 10 cfu) similar results were obtained (data not shown).
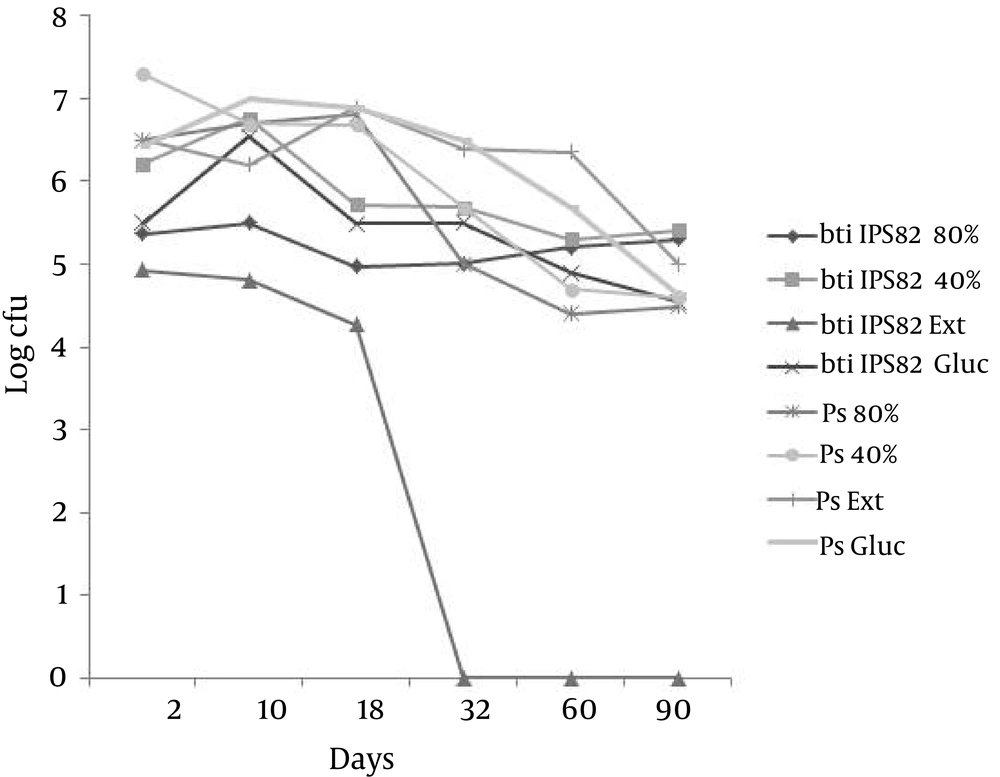
At the C proportion (105: 105 cfu) similar to the A proportion, moisture adjustment had no effect on both bacterial populations yet the addition of yeast extract had an effect on B. thuringiensis population because it disappeared in thirty-two days (Figure 2).
At the E proportion, when the proportion of B. thuringiensis is lower than P. fluorescens (10: 105 cfu), in all treatments, B. thuringiensis population were eliminated during a period of ten to thirty-two days, while populations of P. fluorescens remained present on the ninetieth day (data not shown). In the case of the control proportions, wherein only one of the two populations was inoculated, B. thuringiensis IPS82 survived in order of 1 × 104 cfu until the ninetieth day of the trial while the population of P. fluorescens at the end of the analysis was 1 × 105 cfu (data not shown). These results indicated that the disappearance of B. thuringiensis IPS82 was not dependent on the moisture but the composition of nutrients that could have affected the secretion of toxic compounds into the environment of P. fluorescens or the sensitivity of vegetative cells to the toxic compounds of P. fluorescens secreted in soil.
4.2. Analysis of Sporulation of Bacillus thuringiensis IPS82 in Soil
Bacillus thuringiensis IPS82 is able to transform from a vegetative cell to spore in soil, and vice versa; this transformation could help the survival of the bacteria resisting the antagonism by P. fluorescens. Bacillus thuringiensis IPS82 sporulation percentage in soil was determined on the ninetieth day of the experiment for different inoculated proportions in soil. The results indicated that the recovered cells were mostly spores and not vegetative cells in all proved treatments (Table 1).
| Inoculated Proportions of B. thuringiensis IPS82/P. fluorescens in Soil (cfu) | B. thuringiensis IPS82 Viability on the Ninetieth Day (cfu) | B. thuringiensis IPS82 Sporulation Percentage |
|---|---|---|
| A) 105: 10 | 1.5 × 104 | 100 |
| B) 103: 10 | 3.25 × 104 | 100 |
| C) 105: 105 | 1.22 × 105 | 80 |
| D) 10: 103 | 0 | 0 |
| E) 10: 105 | 0 | 0 |
| F) 103 | 1.87 × 104 | 90 |
Bacillus thuringiensis IPS82 Sporulation Percentage at Different Proportions Inoculated in Soil on the Ninetieth Day
In A and B proportions, 100% of recovered cells, were spores. In D and E proportions, when low amounts of B. thuringiensis were inoculated with respect to P. fluorescens, neither vegetative nor spore cells were recovered.
Information on the reproductive biology and the sporulation process of B. thuringiensis IPS82 in soil is scarce; however, it is possible to speculate that B. thuringiensis IPS82 survives in soil for a brief period of time, as a vegetative cell and it must sporulate for its maintenance in the environment. The sporulation percentage of B. thuringiensis was determined every 24 hours during ten days. The results indicated that recombinant and wild type strains of B. thuringiensis have similar behavior under the same conditions. However, the most interesting result was obtained from the analysis of the onset of sporulation. It was expected for all vegetative cells to sporulate on the first day and remain so, for over ten days (due to low nutrient content in the soil). The results shown in Figure 3, indicate that on the first day, the amount of B. thuringiensis IPS82 obtained from the total population (vegetative cells + spores) and from only the spores, was similar (106 cfu) therefore, all cells were spores, as expected (Figure 3A ). However, from the second day there was a decrease in the number of spores and a slight increase in the number of total cells (Figures 3A and B), indicating that some of the cells in the form of spores germinated and multiplied slightly, maintained a constant cycle of sporulation and germination. This confirms that B. thuringiensis IPS82 similar to Bacillus cereus can germinate, grow and sporulate in soil. The phenomenon of the increment of cells is probably due to the presence of other microorganisms in soil. These microorganisms could segregate some components into the soil, which serve as nutrients for B. thuringiensis development.
4.3. Detection of Bacillus thuringiensis IPS82 and Pseudomonas fluorescens Populations Inoculated in Soil by Molecular Methods
Considering the efficiency of molecular methods, in this study, for the detection of the persistence of these populations, the DNA of B. thuringiensis IPS82 and P. fluorescens populations inoculated in soil was extracted directly from the soil and PCR reaction and dot-blot hybridization were carried out. In the PCR reaction, the Cry 10Aa gene of B. thuringiensis was detected. The design of oligonucleotides for the detection of Cry 10Aa gene was done based on the sequences of three conserved motifs encoding the Cry proteins of B. thuringiensis. The amplification of this fragment was useful and used as a specific probe for monitoring inoculated populations of B. thuringiensis in soil. The PCR reaction showed that the DNA, directly extracted from soil, was suitable to obtain a successful amplification for the Cry 10Aa gene, only when the extracted DNA was diluted (data not shown). This is probably due to some soil components, which could inhibit the reaction, as has been reported in the literature.
In the analysis to detect the inoculated populations of B. thuringiensis IPS82 and P. fluorescens in soil by dot-blot hybridization, the directly extracted DNA from soil was hybridized with a total genomic DNA probe of the analyzed bacteria, B. thuringiensis IPS82 or P. fluorescens. Figure 4 A visualizes the detection of B. thuringiensis IPS82 for hybridization with the B. thuringiensis IPS82 probe of total genomic DNA. The minimal level of detection of this microorganism was 5 × 104 cfu per gram of soil at 10 days, whereas the maximum was 1.5 × 105 cfu per gram of soil, which was found in combination with P. fluorescens. The P. fluorescens detection for hybridization, with the probe of total genomic DNA, from the tenth and the ninetieth day (Figure 4B and C) shows that the minimum level of detection for P. fluorescens was 5 × 103 cfu (on the ninetieth day of the experiment). These results confirm that through genomic hybridization it was possible to monitor the efficiently of the microbial populations introduced in soil, for extended time periods.
5. Discussion
Due to the differences and changes of various environmental conditions at different sites, field studies tend to show large fluctuations in microbial populations (11). Although B. thuringiensis can be isolated from various environments, its role in the ecosystem is not clear. One way to elucidate its role in the ecosystem is studying what happens when these bacteria are introduced into the soil as an ending environment. Earlier, studies showed that the survival of B. thuringiensis IPS82 is affected by interaction with other native soil microorganisms in vitro, particularly, by cohabitation with P. fluorescens (2). Therefore, it is possible to consider B. thuringiensis as a casual soil microorganism, due to its susceptibility to other microorganism antagonistic present in soil (2). In this study the method of counting in plates to follow the inoculated proportions, was an effective tool to estimate the population dynamics and distinguish the change between vegetative cells and spores.
Previous studies have shown that the dormant spores of Bacillus species are much more resistant than their vegetative cell counterparts to a variety of treatments, including heat, pressure, radiation and various chemicals (12). In this study, it was evident that vegetative cells were more sensitive to interaction with other soil microorganisms, in comparison with spores. This study showed that B. thuringiensis, similar to Bacillus cereus, is able to germinate, grow and sporulate in soil (13, 14). A study found a considerable persistence of spores in the soil, yet their number decreased over time; after 135 days from the last application of spores, the number of spores that could be detected was approximately 6.8 log spores g-1 soil (15). This phenomenon is in agreement with the current investigation, wherein was possible to carry out counting of spores in soil with the treatments proved at 90 days of the inoculation.
Other studies showed the persistence of spores of Bacillus sp. in soil for months or years; this data clearly indicates that soil is a favorable reservoir for viable spores, whether the soil is in a rural or urban environment (16, 17). It is also possible that these bacteria use some soil natural components, such as nutrients; according to literature reports, B. thuringiensis BT27a grew in artificial soil with humic acid as the sole carbon and energy source (18). The divergence of the availability of nutrients in different types of soils, explains the difference of isolation of spores in various environments. The exact moment at which the sporulation process was initiated is unknown. In the present investigation two types of populations were inoculated in the vegetative form; B. thuringiensis IPS82 strain and one B. thuringiensis SR08 recombinant strain obtained in the laboratory (9). The B. thuringiensis population sporulation-germination behavior, depended on the availability of nutrients, which could indicate that B. thuringiensis populations fluctuate such as zymogen soil microorganisms do.
At present, culture independent methods based on extraction and analysis of DNA from environmental samples are becoming more popular for assessing the population structure of indigenous or introduced bacterial communities (19-21). In this work molecular methods such as PCR and hybridization using DNA, which was extracted directly from bacterial populations inoculated in soil, were effective to detect and monitor population dynamics, yet PCR was more sensitive to the inhibitor effect of soil components. The literature reports that humic acid, which was present in the soil, could have been interfering with the reaction, inhibiting the activity of enzymes, such as restriction endonucleases and DNA polymerase (8, 22-24). Due to the little information that is available about the B. thuringiensis IPS82 behavior in natural conditions and its interaction with P. fluorescens in soil, this work evaluated the interaction of these populations, using microbiological methods such as traditional culture and molecular methods such as PCR and dot-blot hybridization.
With the purpose of evaluating the population dynamics of B. thuringiensis and its interaction with P. fluorescens under different conditions in soil, the effects of nutrients addition and moisture adjustment were analyzed with the proportions of inoculated bacteria. The results indicated that the disappearance of B. thuringiensis IPS82 was not dependent on the moisture content but the composition of nutrients that could be affecting the secretion of toxic compounds in the environment of P. fluorescens or the sensitivity of vegetative cells to the secreted toxic compounds of P. fluorescens in the soil.
Bacillus thuringiensis IPS82 sporulation percentage in soil was determined on the ninetieth day of the experiment in the inoculated proportions in soil. The results indicated that the recovered cells were mostly spores and not vegetative cells, in all proved treatments. The sporulation percentage of B. thuringiensis IPS82 was determined every 24 hours during ten days. The results indicated that some of the cells in the form of spores germinated and and were increased and maintained in a constant cycle of sporulation and germination. This confirms that B. thuringiensis IPS82 can germinate, grow and sporulate in soil. On the other hand, in this study, for the detection of the persistence of these populations, the DNA of B. thuringiensis IPS82 and P. fluorescens populations inoculated in soil was extracted directly from the soil and the PCR reaction and dot-blot hybridization were carried out. The results confirmed that these methods were efficient for monitoring and detecting the populations inoculated in soil without traditional culture methods.
The data of this work suggests that B. thuringiensis IPS82 can form part of the soil zymogen microorganisms. B. thuringiensis IPS82 can be found in soil in low amounts; its presence in this environment perhaps can be due to soil contamination with this bacterium and the persistence of its spores, so it would be more appropriate to consider this bacterium such as an entomopathogenic microorganism and not as a soil germ due to its ability to kill insects.