1. Background
Until recently, there were only 3 known marine bioluminescent bacterial genera (i.e. Vibrio, Photobacterium, and Shewanella). Nevertheless, a study performed by Urbanczyk et al. in 2007 (1) revealed that the Vibrio fischeri species were different from the other genera of the Vibrionaceae family in terms of phylogenetic and phenotypic properties. Accordingly, V. fischeri and A. logei were reclassified into a new genus called Aliivibrio. In addition to the new classification and according to the results of some studies, the new bioluminescent Vibrio and Photobacterium species were obtained. Up to 2007, only 15 bioluminescent species were known (i.e. V. cholerae, V. fischeri, V. harveyi, A. logei, V. mediterranei, V. orientalis, V. splendidus (biotype I), V. vulnificus, V. salmonicida, Photobacterium angustum, P. leiognathi, P. phosphoreum, P. mandapamensis, Shewanella hanedai, and S. woodyi) (2). Subsequent studies performed in 2007, 2009, and 2010 introduced 6 new bioluminescent species (i.e. P. kishitanii, P. aquimaris, V. azureus, V. sagamiensis,V. chagasii, and A. sifiae) (3-8). There was a special case for V. chagasii because when it was isolated for the first time, it was indicated that this Vibrio species was not bioluminescent (7). A similar situation was demonstrated in a study conducted by Ersoy Omeroglu and Karaboz in 2012 (9). Vibrio gigantis was first isolated by Le Roux et al. in 2005 (10) and is a member of marine bioluminescent bacteria (9). In 2014, this study was cited and it was indicated that the V. gigantis strains were bioluminescent (11). Because of such reasons, the systematic evaluation of bioluminescent bacteria has been regularly revised in terms of their taxonomy, evolutionary relationship, and origin.
The distribution of marine bioluminescent bacteria between all bacteria has yet to be fully elucidated. Marine bioluminescent genera (i.e. Vibrio, Aliivibrio, Photobacterium, and Shewanella) contain a large number of species which are both non-bioluminescent and closely associated with them. Additionally, it is thought that bioluminescent events may have evolved at least 40 times since their onset. Consequently, the isolation and identification of new bioluminescent genera or species from different sources and determination of their genetic diversity are very important. There are different molecular typing methods to investigate the clonal relationships between bacteria. Pulsed-Field Gel Electrophoresis (PFGE) has the highest discriminatory power of all of them. The salient disadvantage of this technique accepted as the gold standard is that the results cannot be compared with each other due to the implementation of dissimilar protocols in different laboratories. In addition, PFGE results in classical protocols as long as three or four days (12). In PFGE, as a consequence of the cutting of the total genomic DNA by restriction endonucleases, PFGE fragment patterns are formed. For a large number of bioluminescent (3, 9, 13-15) and non-bioluminescent bacteria (16), this technique has been used with different restriction enzymes.
2. Objectives
To the best of our knowledge, the existing literature contains no PFGE data performed with the NotI-HF™ restriction enzyme with different bioluminescent bacteria. Thus, in this study, we aimed to determine the genetic relatedness of 10 different bioluminescent species isolated and identified from various marine sources. The data obtained from this study could lead to a better understanding of the bioluminescence mechanism.
3. Materials and Methods
3.1. Bioluminescent Strains
Forty-one bioluminescent strains isolated from different seawater, sediment, and fish samples and collected from regions at discrete depths in the Gulf of Izmir were used for this study. The bioluminescent bacteria identified by phenotypic and molecular methods (9, 15, 17) were streaked onto a Seawater Complete Agar (SWC) medium to obtain a single colony. After checking the purity of the strains, they were grown in a liquid SWC medium (18).
3.2. Preparation of Bioluminescent Strains
Bacterial suspensions with turbidity equivalent to McFarland 1.0 were prepared in 200 μL HST buffer (cell suspension buffer) (10 mM Tris-HCl (Sigma-Aldrich, USA), 50 mM EDTA (Sigma-Aldrich, USA), and 20 mM NaCl (Sigma-Aldrich, USA)), and bacterial pellets were obtained by centrifuging at 13.000 grams for 2 minutes at 4°C. After discarding the supernatants, the pellets were suspended in 200 μL cold HST buffer (12).
3.3. Embedding of Strains into Agarose Plugs
Two percent low-melting agarose (Bio-Rad, USA) was prepared in the HST buffer. The agarose-buffer mixture was supplemented with 0.5 mL SDS (sodium dodecyl sulfate, Sigma-Aldrich; USA) (10 g/50 mL). Afterward, it was melted by heating in a magnetic stirrer at 50°C. The agarose mould (10 mm × 5 mm × 1.5 mm, Bio-Rad, USA) was kept at 4°C. Thereafter, 1.5 mL tubes for each bioluminescent strain were placed into a dry heat block at 50°C and 100 µL of an agarose-SDS mixture. Then, 100 µL of the bacteria suspension in the HST buffer was transferred into them. Subsequently, they were transferred into the agarose moulds without air bubbles and put at 4°C for hardening for 10 minutes (12).
3.4. Fractionation of Bacterial Cell into Agarose
Cell lysis solution I (50 mM Tris-HCl, 50 mM EDTA, 2.5 mg/mL lysosime (Sigma-Aldrich, USA) and 1.5 mg/mL proteinase K (Sigma-Aldrich, USA)) was prepared freshly. Agarose plugs with bacteria were removed from the moulds and placed into the lysis solution. All the tubes were incubated in a water bath at 37°C for 1 hour. After removing the cell lysis solution I, 600 µL of cell lysis solution II was prepared freshly (50 mM EDTA, 1% sarcosyl (Sigma-Aldrich, USA), and 400 µg/mL proteinase K) and was added into the tubes, and they were incubated in a water bath at 55°C for 1 hour (12).
3.5. Washing of Agarose Plugs after Cell Lysis
For solidification, 1.5 mL tubes in a water bath (55°C) were kept in ice for 15 minutes. Then, 5 - 15 mL of sterile ultra-pure water was transferred into each 50 mL of falcon tubes. Afterward, lysis solution II was aspirated and the agarose plugs were transferred carefully with a spatula into these falcon tubes and were incubated in a shaking water bath at 50°C for 15 minutes. Ultra-pure water was fully aspirated. Thereafter, 15 mL of tris-EDTA buffer was transferred into falcon tubes and they were incubated at 50°C with shaking at 15 minutes. This washing step was performed thrice (12).
3.6. Cutting the Whole DNA with the NotI-HF™ Restriction Enzyme in the Agarose Plugs
The agarose plugs containing DNA were cut in the ratio of 1/4 and were placed into a 100 μL fresh enzyme buffer at 37°C for 10 minutes. Next, 100 μL of 1Xbuffer solution was prepared with 100 µm/mL bovine serum albumin (Sigma-Aldrich, USA) and 20 units of fast digest NotI-HF™ (BioLabs 20,000 U/mL, England) and the genomic DNA embedded in agarose plugs were digested with the NotI-HF™ at 37°C for 30 minutes. After incubation, the tubes were held at 4°C for 15 minutes (12).
3.7. Loading the Agarose Plugs into Gel and Electrophoresis
High molar mass restriction fragments were observed by electrophoresis using 1% (w/v) pulsed-field grade agarose (Bio-Rad, USA) prepared with 0.5 X TBE in a CHEF-Mapper PFGE system (Bio-Rad, USA). The separation of the restriction fragments was carried out following running conditions: 6 V/cm2 via 120° angle of impact for 24 hours at 14°C with 5 - 40 seconds pulse times. The gel was stained with ethidium bromide (5 μg/mL, Sigma-Aldrich, USA) and then destained with sterile deionized water (12, 15). The band profile of the bioluminescent strains for the NotI-HF™ restriction enzyme was analyzed using Bio-Profil-1D++ software (Vilber Lourmat, Deutschland) at 10% homology coefficient. Dendrograms belonging to the PFGE band profiles were generated based on the Unweighted Pair-Group Method with Arithmetic Mean (UPGMA) (Vilber Lourmat).
4. Results
Table 1 shows the 10 different bioluminescent strains used in the present study (9).
| Isolation Date of Strains | Isolate | Accession Number | Source | Coordinate | Depth, m |
|---|---|---|---|---|---|
| 30 March 2007 | 0 - 15 | ||||
| V. gigantis | SW15 | JF412215 | Seawater | 38°29’03” N – 26°47’05”E | |
| V. gigantis | SWLiman | JF 412216 | Seawater | 38°27’22” N – 27°09’65”E | |
| A. fischeri | SW22 | JF412240 | Seawater | 38°25’30” N – 26°58’60”E | |
| V. gigantis | SeLu25 | JF412217 | Sediment | 38°23’50” N – 26°39’00”E | |
| 22 January 2008 | 0 - 15 | ||||
| A. fischeri | S2W23 | JF412241 | Seawater | 38°25’09” N – 27°02’86”E | |
| V. gigantis | S2W42 | JF412218 | Seawater | 38°24’58” N – 26°56’88”E | |
| V. gigantis | S2W9 | JF412219 | Seawater | 38°34’99” N – 26°39’00”E | |
| A. logei | Se2Lu45-2 | JF412238 | Sediment | 38°25’03” N – 27°06’09”E | |
| V. gigantis | Se2Lu48 | JF412223 | Sediment | 38°24’75” N – 26°58’90”E | |
| 17 April 2008 | 0 - 15 | ||||
| V. gigantis | S3W46 | JF412220 | Seawater | 38°26’70” N – 27°06’10”E | |
| V. gigantis | S3W28 | JF412221 | Seawater | 38°23’50” N – 26°55’00”E | |
| V. gigantis | S3W2 | JF412222 | Seawater | 38°40’90” N – 26°34’90”E | |
| V. gigantis | Se3Lu25 | JF412224 | Sediment | 38°23’50” N – 26°39’00”E | |
| 06 August 2008 | 0 - 15 | ||||
| V. harveyi | Se4Lu24 | JF412244 | Sediment | 38°26’22” N – 27°04’68”E | |
| V. orientalis | Se4Lu49-2 | JF412251 | Sediment | 38°24’35” N – 26°58’20”E | |
| V. azureus | Se4Lu15 | JF412235 | Sediment | 38°29’03” N – 26°47’05”E | |
| 30 March 2007 | Between 38°37’00”N – 26°42’20”E and 38°37’45”N – 26°43’30”E | 67 | |||
| V. gigantis | FU-10 | JF412225 | Mullus barbatus (internal area) | ||
| V. gigantis | FU-9 | JF412226 | Diplodus annularis (gill) | ||
| V. lentus | FU-7 | JF461265 | Alloteuthis subulata (internal area) | ||
| 30 March 2007 | 50 | ||||
| A. logei | U-6 | JF412239 | Merluccius merluccius (intestine content) | Between 38°32’30” N – 26°45’50”E and 38°31’45”N – 26°45’20”E | |
| P. kishitanii | X-8 | JF412253 | Zeus faber (surface) | Between 38°26’50”N – 26°41’00”E and 38°26’00”N – 26°39’45”E | |
| S. woodyi | X-7 | JF412255 | Pagellus erythrinus (internal area) | ||
| 17 April 2008 | Between 38°34’40”N – 26°46’10”E and 38°33’45”N-26°46’55”E | 42 - 44 | |||
| V. gigantis | E-14 | JF412227 | Lepidotrigla cavillone (gill) | ||
| V. gigantis | E-16 | JF412228 | Boops boops (surface) | ||
| V. gigantis | E-15 | JF412229 | D. annularis (surface) | ||
| V. gigantis | E-10 | JF412230 | Citharus linguatula (gill) | ||
| V. gigantis | E-11 | JF412231 | Arnoglossus laterna (internal area) | ||
| A. fischeri | E-4 | JF412242 | P. erythrinus (gill) | ||
| 06 August 2008 | Between 38°34’40”N – 26°46’10”E and 38°33’45”N-26°46’55”E | 42 - 44 | |||
| V. harveyi | H-15 | JF412245 | C. linguatula (internal area) | ||
| V. harveyi | H-5 | JF412246 | Scorpaena notata (internal area) | ||
| V. azureus | H-1 | JF412236 | D. vulgaris (gill) | ||
| V. azureus | H-14 | JF412237 | Trisopterus minutuscapelanus (surface) | ||
| V. harveyi | H-11 | JF412247 | P. bogaraveo (surface) | ||
| V. crassostreae | H-7 | JF412248 | A. laterna (gill) | ||
| V. gigantis | H-3 | JF412232 | M. merluccius (surface) | ||
| V. crassostreae | H-12 | JF412249 | Engraulis encrasicolus (gill) | ||
| V. crassostreae | H-19 | JF412250 | Conger conger (internal area) | ||
| A. fischeri | H-18 | JF412243 | Dentex macrophthalmus (internal area) | ||
| V. gigantis | H-2 | JF412233 | D. annularis (intestine contents) | ||
| V. gigantis | H-16 | JF412234 | B. boops (gill) | ||
| V. orientalis | H-9 | JF412252 | L. cavillone (gill) |
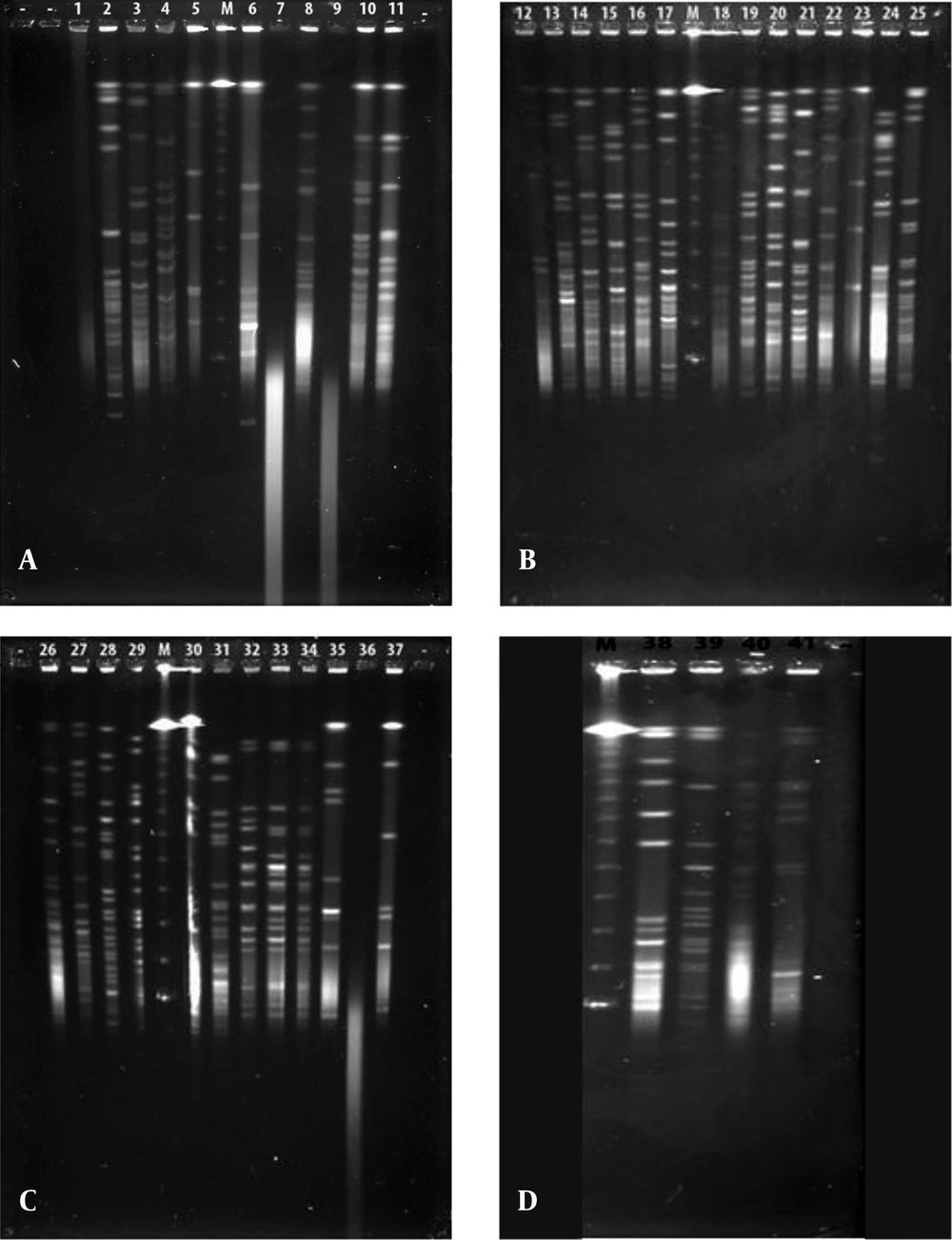
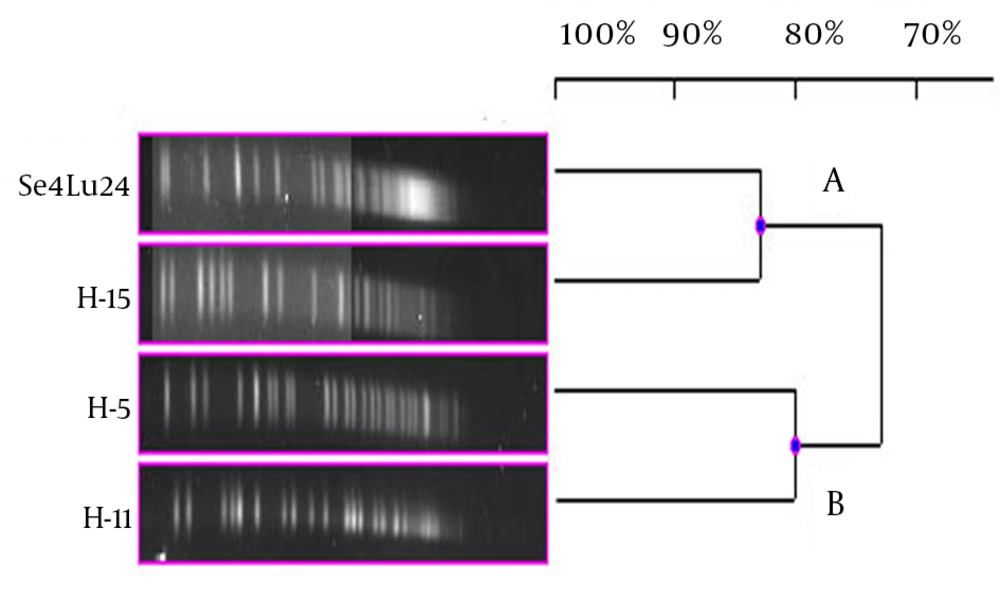
Each different group was evaluated separately in terms of PFGE fragment patterns. Three groups were composed (i.e. V. harveyi, V. gigantis, and other species). As is seen in Figure 1, all the V. harveyi strains could be typed with PFGE performed with the NotI-HF™ restriction enzyme (Figure 1 C). As a result of the NotI-HF™ digestion, 2 main clusters (A and B) with 73% pattern homology were obtained. The closest strains with respect to genetic resemblance were achieved (i.e. Se4Lu24 and H-15 strains) with 83% homology. The other strains (i.e. H-5 and H-11) demonstrated 80% pattern homology (Figure 2).
A, V. gigantis (1:E-16, 2:E-10, 3:E-11, 4:H-3, 7:S3W46, 8:S3W28, 9:H-16, 10:E-15, 11:H-2, 25:E-14), A. logei (5:Se2Lu45-2), and A. fischeri (6:S2W23) strains; B, V. gigantis (12:SW15, 13:SWLiman , 14:SeLu25 , 15:S2W42 , 16:S2W9 , 17:S3W2, 18:Se2Lu48 , 19:Se3Lu25, 20:FU-10, 21:FU-9), V. lentus (22:FU-7), A. logei (23:U-6), and S. woodyi (24:X-7) strains; C, V. harveyi (26:Se4Lu24, 27:H-15, 28:H-5, 29:H-11), V. orientalis (30:Se4Lu49-2, 31:H-9), V. crassostreae (32:H-7, 33:H-12, 34:H-19), and A. fischeri (35:SW22, 36:E-4, 37:H-18); and D, V. azureus (38:Se4Lu15, 39:H-1, 40:H-14) and P. kishitanii (41:X-8), M: Marker (Lambda Leader, Bio-Rad, USA, 48,5 kb - 1,000 kb).
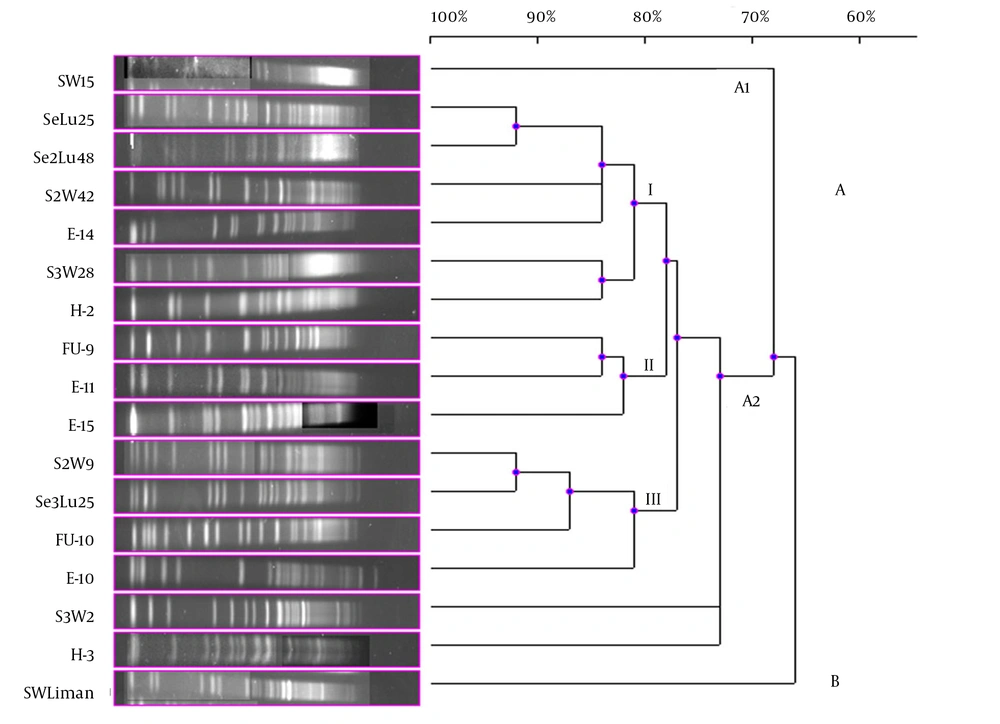
Although 20 V. gigantis strains were used in this study, only 17 strains were typed with PFGE performed with the NotI-HF™ restriction enzyme. As a result of the PFGE analysis for the V. gigantis strains, 17 different PFGE fragment patterns were obtained (Figure 1 A and B). PFGE with the NotI-HF™ restriction enzyme resulted in 2 main clusters (A and B) and 17 different PFGE fragment patterns. V. gigantis strains S3W46, E-16, and H-16 could not be typed. Clusters A and B exhibited 66% pattern homology. Cluster A consisted of two subclusters (A1 and A2), which displayed 68% pattern homology, with 16 different PFGE patterns that showed pattern homology between themselves ranging from 68 to 92%. In all the V. gigantis strains, the unrelated strain was SWLiman with 66% pattern homology. It was determined that the closest strains in terms of PFGE fragment patterns were observed between the SeLu25 and Se2Lu48 strains isolated from sediment and S2W9 and Se3Lu25 with 92% homology. In subcluster A2, 3 different groups (I, II, and III) were formed with 73% pattern homology. Thirteen different band patterns were achieved in group I with pattern homology between 81% and 92% (Figure 3).
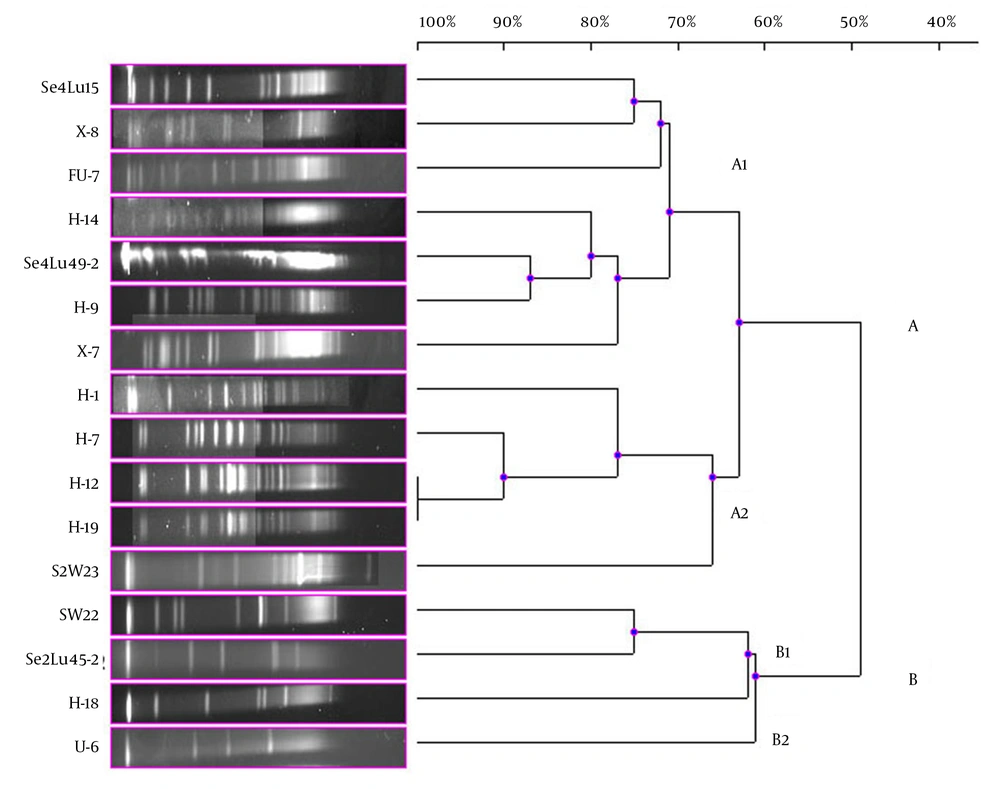
In the current study, 41 bioluminescent strains isolated and identified were used and the remaining 17 strains were evaluated together from the point of PFGE fragment patterns with the NotI-HF™. However, only 16 strains were typed by this technique (Figure 1). The PFGE band pattern for the A. fischeri strain E-4 could not be achieved. In this group, there were 3 V. azureus strains, 3 V. crassostreae strains, 2 V. orientalis strains, 3 A. fischeri strains, 2 A. logei strains, 1 P. kishitanii strain, 1 V. lentus strain, and 1 S. woodyi strain. In consequence of the NotI-HF™ digestion of the total genomic DNA, 2 main clusters (A and B) formed with 49% pattern homology. Considering all the bioluminescent strains in this group, PFGE fragment patterns with genome homology between 61% and 100% were achieved. Two V. crassostreae strains (i.e. H-12 and H-9) exhibited genome homology with the highest percentage (100%). Also, it was determined that the other V. crassostreae strain (i.e. H-7) was similar to the H-12 and H-9 strains at a ratio of 90% pattern homology. While the V. orientalis strains Se4Lu49-2 and H-9 had 87% genome homology, the A. fischeri (S2W23, SW22, and H-18) and A. logei (Se2Lu45-2 and U-6) strains between each other showed the lowest homology (61% and 62%). With the exception of S2W23, all the A. strains were located in cluster A (Figure 4).
Dendrogram, Showing the Genetic Relationships Between the Bioluminescent V. lentus, V. azureus, V. orientalis, S. woodyi, V. crassostreae, A. fischeri, A. logei, and P. kishitanii Strains Based on the Pulsed-Field Gel Electrophoresis Analysis of the Genomic Restriction Fragments Formed with the NotI-HF™
5. Discussion
Within the scope of our study, the genetic diversity of 41 bioluminescent strains was determined. Ten different bioluminescent species were evaluated in terms of genomic polymorphism based on PFGE with the NotI-HF™ restriction enzyme. Moreover, so far, there have been no data indicating that the V. lentus and V. crassostreae strains are bioluminescent. It is, therefore, believed that these data will contribute to a better understanding of the evolution of bioluminescence. There are a large number of techniques for the characterization of bacterial species and also the evaluation of DNA similarity between two bacterial strains. The first definition method was DNA-DNA hybridization, but it is not suitable for the routine identification or determination of the genetic polymorphism of isolates because, although it is necessary to exhibit new bacterial genera and species, this technique is complex and limited to a few laboratories and also there is a dearth of relevant information (19).
To investigate clonal diversity and relationship between bacteria, PFGE with the highest discriminatory power has been used (12). PFGE is considered the gold standard because the structural integrity of the chromosome of the bacteria embedded in agarose plugs is not destroyed and the whole genome is profiled via cutting with the restriction enzyme. It has been proven that PFGE genomic DNA analysis is more discriminative in exhibiting phylogenetic diversity than analyses based on phenotype characterization (20). The genetic diversity and genotyping of some bioluminescent Vibrio isolates were determined by cutting their whole genome with 5 different restriction enzymes (i.e. ApaI, EagI, NotI-HF™, SpeI, and SmaI). DNA profiling with PFGE was introduced by Suwanto et al. in 1998 (13). In addition, some other studies have revealed further data relating to the PFGE analysis of bioluminescent bacteria (3, 21). In the present study, we determined clonal diversity and relationship using PFGE among bioluminescent S. woodyi strains (15) and V. gigantis strains (9). Nonetheless, to the best of our knowledge, the existing literature lacks research on the determination of genetic diversity via PFGE among bioluminescent A. fischeri, A. logei, V. azureus, V. orientalis, V. lentus, and V. crassostreae strains. The current study is, therefore, the first of its kind to report this genetic diversity. It is also deserving of note that the PFGE analysis of the V. gigantis strains with the NotI-HF™ has been carried out for the first time in the present study. The findings are very significant since V. gigantis was first reported as a bioluminescent strain in one of our previous studies (9). For assays, the NotI-HF™ was selected in light of a study performed by Suwanto et al. (13). The selection of the restriction enzyme is a critical variable in the PFGE process. Clonal diversity and relationship among the isolates are determined based on the restriction fragment band pattern. Accordingly, the fragments should be workable in number and distribution.
In the current study, 4 out of the 41 bioluminescent strains could not be typed by PFGE with the NotI-HF™ and although optimization assays were made and repeated three times for all the PFGE experiments, a smear appeared. Similarly, in a study performed by Eddabra et al. (21), 5 of the 30 Vibrio isolates could not be typed by PFGE with the NotI-HF™ due to the methylation of genomic DNA or DNA degradation during the process. In our study, because all the bioluminescent strains were isolated from different sources and depths, our findings reconfirm the heterogeneous structure of bioluminescent strains (21). In consequence of PFGE assays, 17 different band patterns were obtained as was expected. These differences obtained were not determined by phenotypic characterization. Whereas our previous study had determined that the bioluminescent V. gigantis strains had a few differences in their phenotypes, the findings of the present study revealed that the V. gigantis strains had a high incidence of polymorphism when the whole genome was analyzed via PFGE with the NotI-HF™.
The highest percentage of genome similarity in the 17 V. gigantis strains used in the current experiment was found to be 92%. SeLu25 and Se2Lu48 were two sediment strains isolated from different seasons and stations but from the same depth. Additionally, S2W9 and Se3Lu25 were seawater and sediment strains, respectively, and even though they were isolated from the same depth, their isolation sources were different. In contrast, the SeLu25-Se2Lu48 (92%) and S2W9-Se3Lu25 (92%) strains were found to be genetically closer than H-2, FU-9, and E-15 (78%) isolated from the same fish species. Although SW15, SWLiman, and SeLu25 were isolated from the same depth in the same season, it was become evident that SWLiman was different from all other V. gigantis strains and it showed 66% genome similarity with the others. In addition, it was determined that only bioluminescent V. gigantis strains showed distribution in Diplodus annularis and they were not completely identical in terms of whole genome and also genotypic similarity among them was at a ratio of 78%.
As a result of the PFGE assays, all the V. harveyi strains were typed and 4 different PFGE band patterns were achieved. As well as being isolated in the same season; while sediment sample Se4Lu24 was obtained from a depth of 0 - 15 meters, the other V. harveyi strains (i.e. H-15, H-5, and H-11) were taken from a depth of 42 - 44 meters. In consequence of the PFGE assay, these strains were divided into 2 main clusters with 73% of pattern homology. Contrary to expectation, the closest genome similarity (83%) was achieved from 2 strains, Se4Lu24 (a sediment isolate) and H-15 (isolated from Citharus linguatula). To determine why bioluminescence has come into being in the evolutionary process, some studies have been performed, especially on the V. harveyi strains, and significant results have been obtained (22). Hence, the determination of the genetic proximity of these strains to one another will contribute greatly to the understanding of these mechanisms and we hope that our findings will play a part in future investigations.
In the present study, the remaining 17 bioluminescent strains were examined together. A. fischeriE-4 strain was not typed. On dendrogram consisting of the 2 main clusters, it was observed that the Aliivibrio strains were gathered in cluster B and the other bioluminescent strains were in cluster A. Then again, it was determined that the Aliivibrio strains were different from the other bioluminescent Aliivibrio and Photobacterium species and their genome similarity was 49%. Only the A. fischeriS2W23 strain was located in cluster A and it showed 63% of genome similarity with these species in a different subcluster of cluster A. The Aliivibrio genus was incorporated in the Vibrionaceae family in 2007 in the sequel of a study performed by Urbanczyk et al. (1), who reported that some Vibrio species were different from the species in the Vibrionaceae family in terms of phylogenetic and phenotypic features. These results chime in with the findings of our study.
A similar situation was observed in the bioluminescent strain V. azureus. The results showed that 3 V. azureus strains emerged in different subclusters in cluster A and each of them displayed higher genome homology with different bioluminescent strains than similarity among themselves. Contrary to this, a high PFGE band pattern resemblance was determined in the V. orientalis and V. crassostreae strains. Based on the band pattern, especially 2 V. crassostreae strains (i.e. H-12 and H-19) were completely identical and the other V. crassostreae strain (i.e. H-7) exhibited 90% genome homology with these 2 strains. The V. orientalis strains Se4Lu49-2 and H-9 were more similar to each other in terms of the genomic feature than the other species, and the homology of the band pattern was 87%.
The V. lentus, P. kishitanii, and V. azureus strains, which were first reported in the literature in 2001, 2007, and 2010, respectively, were located in the same subcluster in the present study. Our results showed that especially the new bioluminescent strains, i.e. P. kishitanii and V. azureus, had 75% of the PFGE band pattern homology. The V. lentus strains were isolated and identified for the first time in 2001 and characterized by various research groups in subsequent years (23, 24). However, none of these studies showed that this strain was bioluminescent. Our findings revealed that the V. lentus strains could also show bioluminescence and had genome similarity at a ratio of 75% with the bioluminescent species recently reported in the literature. In addition, the PFGE analysis demonstrated that the V. lentus strains emerged separately from the other bioluminescent strains in the different subclusters.
The PFGE genomic DNA analysis is more discriminative in revealing genetic diversity and polymorphism among related species than analyses based on phenotypic characterization. A study performed by Suwanto (20) revealed that 25 isolates characterized by physiological analysis came together under 2 Vibrio species. Moreover, when the PFGE analysis with the NotI-HF™ was performed, 13 different genotypes were ingenerated. The analyses of the phenotypic characterization of these strains have verified that the discriminatory power of PFGE is higher than that of the other techniques. Our results revealed that 37 bioluminescent strains belonged to 4 different genera and 10 different species had 36 different genotypes.
To our knowledge, there have not been any PFGE studies including 4 bioluminescent genera. The present study is, therefore, the first of its kind in terms of its scope. With the discovery of new bioluminescent species exhibiting new bioluminescent features, we hope that the findings of the present study will contribute to a better understanding of the mechanism and evolution of bioluminescence and polymorphism in different bioluminescent strains. The strains investigated in the current study were isolated and identified from different marine sources and consisted of the 4 bioluminescent genera identified so far. Moreover, in the present study, the PFGE assay was performed by using the NotI-HF™ restriction enzyme and the genetic diversity of 37 bioluminescent strains was demonstrated.