1. Background
Staphylococcus aureus, as a harmful pathogen, is able to express numerous virulence factors. Beside the enterotoxin superantigens, S. aureus exerts pathogenicity through producing some pyrogenic toxins such as the tsst-1 gene (1). The tsst-1 gene encodes a 21.9 KDa extracellular toxin causing toxic shock syndrome (TSS) (2). It is known as a severe acute disease distinguished by symptoms such as fever, rash, hypotension and dysfunction of multiorgan systems. In addition, TSS secretion into the human blood may raise the rate of neonatal TSS-like exanthematous disease, Kawasaki syndrome and sudden infant death syndrome (3). Although the frequency of the tsst-1 gene within methicillin-sensitive Staphylococcus aureus strains (MSSA) is already high, recently, the number of MRSA strains harboring this gene has increased, reportedly (4-6). The last findings have reported the correlation between the incidence of TSS and tampon use by women, because of the colonization of strains that produce toxic shock syndrome toxin (TSST) in the vagina. However, to date, this serious illness may occur in various conditions such as a surgical complication or localized infections at other sites such as skin or respiratory tracts (1).
Choice antibiotics for treating S. aureus are methicillin and vancomycin. Deficiency of appropriate therapy may cause a lethal shock during 24 hours after the first symptom manifestations (1). Therefore, having information about the frequency of this gene and its relation to antibiotic resistant patterns might be helpful in limiting the death rate, in due course.
To our knowledge, there are no findings on the prevalence of TSST-producing S. aureus strains isolated from clinical samples in Iran. In this study, we aimed to investigate the distribution of S. aureus strains carrying the tsst-1 gene in the clinical isolates. In addition, the existence of mecA gene and the antimicrobial susceptibility pattern of each strain and their association with each other were determined.
2. Objectives
The purpose of the current study is to evaluate the incidence of the toxic shock syndrome toxin (tsst-1) gene and its association with the prevalence of the mecA gene and drug resistance.
3. Materials and Methods
3.1. Bacterial Isolates
The sum total of 197 S. aureus strains were randomly collected from clinical samples including wounds, blood, bile, tracheal aspirate, sputum, ear, chest tube, bronchoalveolar lavage (BAL), bone, urine, stool, synovial and pleural effusions during the years 2011 and 2012 from one of the educational hospitals in northern Tehran, Iran. The isolated S. aureus strains were confirmed using catalase, coagulase and DNase activity tests, a gram-staining test, mannitol fermentation and the presence of hemolysis on the blood agar plate after 24 hours growth at 37°C. Standard S. aureus strains named MN8 and ATCC 25923 were applied as positive and negative controls, respectively.
3.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility status toward 12 antibiotics oxacillin (1 µg), penicillin G (10 un), vancomycin (30 µg), ciprofloxacin (5 µg), erythromycin (15 µg), clindamycin (2 µg), cotrimoxazole (25 µg), gentamicin (10 mg), linezolid (30 mg), cephazolin (30 mg), kanamycin (30 mg) and tetracycline (30 µg) were evaluated for each isolate. The evaluation was carried out by means of disc diffusion method (Kirby-Bauer) according to clinical and laboratory standard institute (CLSI) guidelines (7). All antibiotics were produced by Mast, England.
3.3. Primer Designing
Primers were designed based on the sequences of the tsst-1 and mecA genes of S. aureus extracted from Gene Bank, applying Gene Runner programs. To evaluate the specificity of designed primers, they were analyzed against Primer-BLAST. The sequences of the primers, synthesized by SinaClon Co, Iran, were as follows:
tsst-1 forward 5’CTGGTATAGTAGTGGGTCTG3’,
tsst-1 reverse 5’AGGTAGTTCTATTGGAGTAGG3’,
mecA forward 5’-TGAGTTGAACCTGGTGAAGTT-3’ and
mecA reverse 5’-TGGTATGTGGAAGTTAGATTGG-3’.
For further corroboration, the PCR product of each gene was analyzed by sequencing (Macrogen Research, Seoul, Korea).
3.4. DNA Isolation and PCR Amplification for Gene Detection
The PCR method was used to investigate the distribution of the tsst-1 and mecA genes among the isolates. Briefly, each isolate was cultured in 5 mL of LB broth (Merck, Germany) at 37°C overnight under aerobic conditions, and then bacterial cells were harvested by centrifugation at 6000 rpm for 10 minutes in 100 μL buffer solution (10 mM Tris HCl, pH 8.0; 1 mM EDTA; 100 mM NaCl), and 15 μL lysozyme (0.25 mg/mL) (SinaClon, Iran) were added to a bacterial pellet and incubated for an hour at 37°C. After that, 200 μL GC solution and 15μL proteinase K solution (Sigma-Aldrich, Germany) were added and incubated for 15 minutes at 60°C. Finally, the DNA was precipitated using isopropanol (Merck, Germany). The quality and quantity of purified DNA were investigated using electrophoresis and a NanoDrop spectrophotometer (NanoDrop 1000, Wilmington, DE), respectively. One µL of each DNA was amplified in 25 μL of mixture reaction consisting of a 10X reaction buffer, 0.1 mM of the deoxynucleoside triphosphates (dNTPs), 2 mM MgCl2, 10 ρmol of designed primers and 1 U of Taq DNA polymerase (SinaClon, Iran). PCR was performed by Eppendorf asterCycker (Hamburg, Germany) with an initial denaturation step of 5 minutes at 94°C; 35 cycles of 1 minute at 94°C; 2 minutes at 54°C; 50.9°C and 57°C as an annealing temperature for the tsst-1 and mecA genes, respectively; and 1 minute at 72°C followed by a 5 minutes final extension at 72°C. For determining the presence of the desired amplicon, electrophoresis was done on 1.5% gel agarose stained by ethidium bromide, and then the products were visualized by UV transilluminator (Kiagene, Iran). The sizes of the PCR product of amplifying the tsst-1 and mecA genes were 271 bp and 855 bp, respectively.
3.5. Evaluation of the Sensitivity and the Specificity of PCR
DNA isolated from S. epidermidis, S. saprophyticus, Enterococcus faecalis, Clostridium perfringens and C. difficile was tested to assess the specificity of the designed primers. Moreover, the sensitivity of PCR tests was determined by preparing two-fold serial dilutions of the genomic DNA purified from S. aureus in the range of 1 ng to 32 ng. The PCR procedure was performed for each analysis under the protocol described before.
3.6. Statistical Analyses
To compute the correlation between the incidence of mecA and tsst-1 genes, drug resistance and the demographic properties of patients, we performed the chi-square test and Fisher’s exact test using the SPSS version 15 software package (Chicago, IL). A P ˂ 0.05 was considered statistically significant.
4. Results
4.1. Strain Isolates
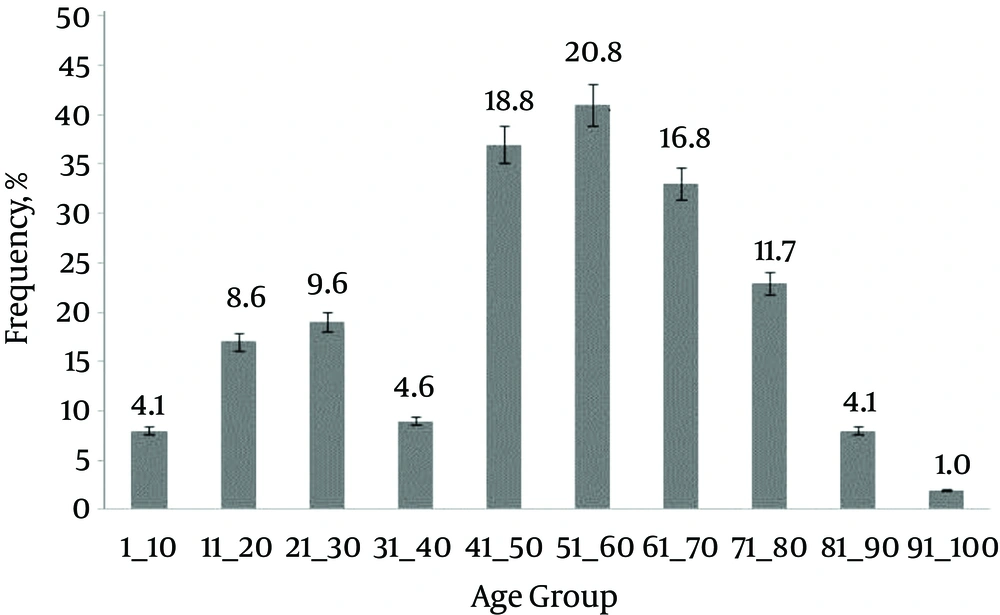
In this study, we evaluated the prevalence in the clinical isolates of S. aureus that contained the tsst-1 gene. Out of 197 strains, 85 (43.1%) were from wounds, 18 (9.13%) from blood, nine (4.56%) from bile, 70 (35.5%) from tracheal aspirate, six (3.1%) from sputum and two (1%) were from ear samples. One strain was separated from each one of the chest tubes (0.5%), BAL (0.5%), bone (0.5%), urine (0.5%), stool (0.5%), pleural (0.5%) and synovial samples (0.5%). Among the isolates, 136 (69%) were from males and the rest (31%) were from females. The ages of our cases were in the range of 1 to 94 years, categorized in 10 groups. Most cases belonged to the age group of 51 to 60 years. Figure 1 shows the frequency of the cases with respect to age range.
4.2. Distribution of the tsst-1 and mecA Genes in the Clinical Isolates
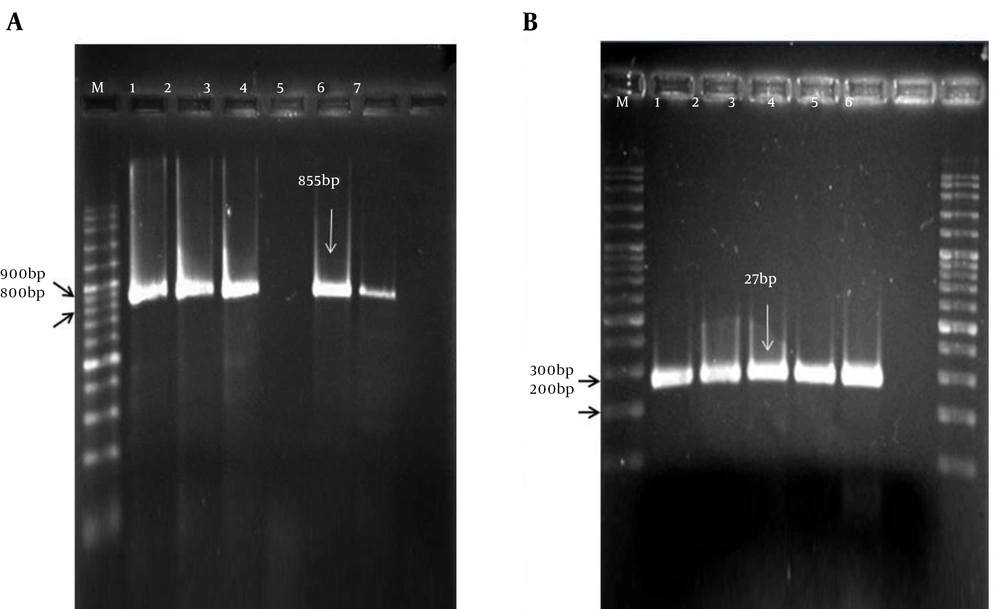
The frequency of the two genes was identified using PCR, and their relation to gender, age and sample type was investigated. Representative examples of PCR reactions for the tsst-1 and mecA gene identification are shown in Figure 2. To test the specificity of the designed primers, we tested DNA purified from S. epidermidis, S. saprophyticus, E. faecalis, C. perfringens and C. difficile. No PCR product was slighted (data not shown). Furthermore, results from the sensitivity analysis showed the primer for the tsst-1 gene was able to identify S. aureus with a minimum concentration of 2 ng.
A, The amplicon size of mecA gene is 855 bp; M indicates the 100 bp DNA ladder; lane 1 is a positive control (S. aureus ATCC 43300); lanes 2 - 6 indicate results from clinical isolates; and lane 7 is a negative control (S. aureus ATCC 25923); B, The PCR product of tsst-1 gene is 271 bp; M, 100 bp DNA ladder; lane 1 is a positive control; lanes 2 - 5 are clinical isolates; and lane 6, a negative control.
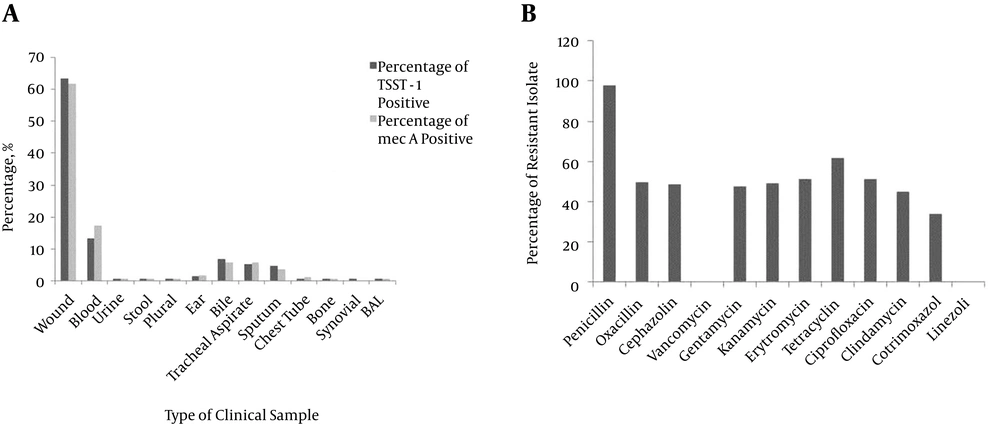
Of the 197 isolates, 134 (68%) and 172 (87.3%) possessed the tsst-1 and mecA genes, respectively. Figure 3 A shows the distribution of the mecA gene in each type of isolate. The highest amount of mecA positive isolates were separated from the wound (61.2%) and blood (17.6%) samples. Moreover, no mecA gene was detected in the synovial sample. The mecA gene was present in 115 (84.5%) of the males and 55 (90.1%) of the females. No significant differences were seen between the sample type, gender and age with the existence of the mecA gene (P = 0.3, 0.7 and 0.054, respectively) (Figure 3).
Among the tsst-1 positive strains, 85 were obtained from wounds (63. 4%), 18 from blood (13. 4%), nine from bile (6. 7%), seven from tracheal aspirate (5. 2%), six from sputum (4. 5%) and two from (%2) ear samples. All strains isolated from chest tube, BAL, bone, urine, stool, pleural and synovial samples had the tsst-1 gene (Figure 3 A). The tsst-1 gene positive strains occurred more often in male cases. Strains separated from 93 (47.2%) male and 41 (20.8%) female samples contained the tsst-1 gene. No statistically significant discrepancy was seen between the tsst-1 gene with respect to gender, age and the source of clinical samples.
The tsst-1 gene was found in 120 (69.8%) of 172 MRSA cases. In addition, this gene was detected within 14 (56%) of MSSA isolates. However, we found no significant correlation between the carriage of the two mentioned genes among isolates (P = 0.126).
4.3. The Antimicrobial Susceptibility Analysis
To determine the antibiotic resistant properties of each isolate, an antimicrobial susceptibility analysis was done. The highest resistance was to penicillin. No isolate was resistant to linezolid and vancomycin. Figure 3 B shows the frequency of resistant isolates to each antibiotic. Multi-drug resistance (resistance to more than three antibiotics). Status of our isolates is summarized in Table 1. Data from the statistical analysis showed no significant impact by sex, age and type of samples on the incidence of multi-drug resistance properties (P = 0.794, 0.526 and 0.888, respectively).
| Number of Antibiotic Resistance | mecA Geneb | tsst-1 Geneb | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| 1 | 49 (83.1) | 10 (16.9) | 40 (67.8) | 19 (32.2) |
| 2 | 23 (69.7) | 10 (30.3) | 23 (66.7) | 10 (30.3) |
| 3 | 6 (100) | 0 (0) | 5 (83.3) | 1 (16.7) |
| 4 | 2 (100) | 0 (0) | 2 (100) | 0 (0) |
| 5 | 1 (100) | 0 (0) | 0 (0) | 1 (100) |
| 7 | 10 (100) | 0 (0) | 6 (60) | 4 (40) |
| 8 | 1 (100) | 0 (0) | 0 (0) | 1 (100) |
| 9 | 22 (91.7) | 2 (8.3) | 14 (58.3) | 10 (41.7) |
| 10 | 58 (95.1) | 3 (4.9) | 44 (72.1) | 17 (27.9) |
| Total | 172 (100) | 25 (100) | 134 (100) | 63 (100) |
aValues are expressed as No. (%).
bP value is 0.032 for mecA and 0.4 for tsst-1.
Results from assessing the prevalence of mecA gene and the antibiotic resistance status of isolates are given in Table 2. Except for penicillin, ciprofloxacin, vancomycin and linezolid, resistance to other tested antibiotics rises significantly with the presence of the mecA gene (Table 2). Moreover, we observed that the presence of the mecA gene increases significantly among the multi-drug resistance isolates (P = 0.032) (Table 1). Fifty-eight (33.7%) of MRSA isolates were resistant to all antibiotics except vancomycin and linezolid.
| Gene Antibiotic Resistance | mecA | tsst-1 | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | P Value | Positive | Negative | P Value | |
| Penicillin | 169 (98.3) | 24 (96) | 0.422 | 132 (98.5) | 61 (96.8) | 0.383 |
| Oxacillin | 92 (53.5) | 6 (24) | 0.005 | 65 (48.5) | 33 (52.4) | 0.362 |
| Cephazolin | 91 (52.9) | 5 (20) | 0.002 | 62(46.3) | 34 (54) | 0.196 |
| Vancomycin | 0 (0) | 0 (0) | - | 0 (0) | 0 (0) | - |
| Gentamicin | 89 (51.7) | 5 (20) | 0.002 | 63 (47) | 31 (44.2) | 0.446 |
| Kanamycin | 91 (52.9) | 6 (24) | 0.006 | 65 (48.5) | 32 (50.8) | 0.442 |
| Erythromycin | 95 (55.2) | 6 (24) | 0.003 | 68 (50.7) | 33 (52.4) | 0.476 |
| Tetracycline | 111 (64.5) | 10 (40) | 0.017 | 84 (62.7) | 37 (58.7) | 0.352 |
| Ciprofloxacin | 92 (53.5) | 9 (36) | 0.077 | 67 (50) | 34 (54) | 0.357 |
| Clindamycin | 83 (48.3) | 5 (20) | 0.006 | 59 (44) | 29 (46) | 0.455 |
| Cotrimoxazole | 63 (36.6) | 4(16) | 0.031 | 47 (35.1) | 20 (31.7) | 0.385 |
| Linezolid | 0 | 0 | - | 0 | 0 | - |
| Total | 172 (100) | 25 (100) | - | 134 (100) | 63 (100) | - |
aValues are expressed as No.(%).
Forty-four (32.8%) of isolates harboring the tsst-1 gene were resistant to all described antibiotics except vancomycin and linezolid (Table 2). However, the antibiotic resistance pattern of strains without the tsst-1 gene was similar. There is no significant correlation between the distribution of tsst-1 gene and the multi-drug resistance pattern (P = 0.4) (Table 1).
5. Discussion
Staphylococcus aureus causes high rate of morbidity and mortality because of severe nosocomial infections (8). The most important issue is difficulty in treatment due to emerging MRSA strains and its increasing prevalence in some parts of the world. Besides, multi-drug resistant properties of MRSA strains and the production of various types of virulence factors let the infections spread easily. It is believed that resistance to antibiotics can alter the expression of genes involving in the pathogenesis (9).
Here, the genotyping outcomes demonstrated an increased incidence of MRSA strains (87.6%) with a high rate of antibiotic resistance; these are similar to findings in several reports from China (77.65% - 80%) (10, 11). Nevertheless, findings from most studies conflict with the current study. The prevalence of MRSA that has been observed is 0.3% - 34% in European countries (12-14), 41.6% in the USA (15), 16.2% in Nigeria (16) and 18% in Russia (17).
In a study by Tokajian et al., 72% of isolates in Lebanon were MRSA, but a lower incidence of multi-drug resistance properties (18%) has been reported (11). Differences in the pattern of mecA gene distribution with regard to gender and age were not obvious in both MRSA and MSSA groups. However, we found this gene more prevalent among the wound samples. The frequency of mecA gene has been reported to be 63% by Rahimi et al. and 58% by Shahkarami et al. (18, 19).
The prevalence of tsst-1 gene in our isolates was more abundant (68%) than in previous studies. The frequency of this gene has been reported as less than 20% by El-Ghodban et al. (20), Tsen et al. (21), Demir et al. (22), Liu et al. (23), Yu et al. (15), Becker et al. (24), Fenner et al. (25), Hoseini Alfatemi et al. (26) and Schlebusch et al. (27). In a study performed by Xie et al and Kimura et al. (28, 29), this amount was 48.1%. The frequency of the tsst-1 gene in our isolates was higher than in findings from the literature (30, 31). Most of the previous studies evaluated the presence of a mentioned gene at a protein level (23, 24, 32). On the other hand, the presence of gene does not mean the expression of protein. Since none of our patients had TSS, it could be inferred that the toxin did not express among the tested population.
In contrast to results from the early studies reporting a high prevalence of tsst-1 among MSSA strains (11, 15, 25, 27, 32), several recent researchers observed a high rate of tsst-1 in MRSA strains (23, 33). Although 72.6% of our isolates were MRSA, the presence or absence of tsst-1 gene was not statistically significant between both MSSA and MRSA groups.
Nowadays, S. aureus strains resistant to numerous antimicrobial drugs are commonly isolated from nosocomial infections. The spreading of multi-drug resistant strains is one of the major challenges in healthcare all around the world, owing to difficult treatment. In a retrospective study by Mohaghegh et al. in Iran from 2006 to 2011, antibiotic resistance increased among clinical S. aureus strains (34). In this study, the frequency of antimicrobial resistance was very high. Our results showed a high rate of resistance to many antibiotics that are routinely administered for treating the staphylococcal infection. Similar to most regions of the world (15-17), resistance to penicillin was remarkably high in our studied population (94.4%). Despite the high frequency of MRSA among our isolates, half of the specimens were resistant to oxacillin, which was lower than in records from Belgium (99%) (35) and higher than in those from Lebanon (32%) (11) and Nigeria (16.2%) (16). Accordingly, using the screening test based on the oxacillin resistance assay might lose some MRSA data. Inconsistent with data from some Asian and African countries in which the level of resistance to erythromycin was lower than 30% (36, 37) or was 0% (16, 17), erythromycin resistant strains were more abundant in the current study (51.3%) but lower than in the United Kingdom (90%) (38), China (97.8%) (23) and Australia (98%) (39). We also observed an increased prevalence of tetracycline resistant strains (61.3%) compared to previous reports from Lebanon (48% and 44%) (11, 40) and the USA (5%) (41). However, Nimmo et al. and Zhang et al. reported the prevalence of 80% and 97.8% resistant isolates from Australia (39) and China (23), respectively. Moreover, the incidence of gentamicin resistant strains was higher (47.7%) than in records from Nigeria (14.7%) (16), China (28.1%) (15) and Russia (19%) (17). Also, the rate of resistance to clindamycin and ciprofloxacin was higher than those from Russia (17), Nigeria (16), Libya (20) and Lebanon (11). Similar to results from several previous studies, resistance to vancomycin (11, 37, 38, 40, 42) and linezolid (15, 16) was not seen among our isolates.
Three important findings were attained through the current study. First, the percentage of MRSA strains and consequent multi-drug resistance has risen in our population. Second, despite the increasing incidence of tsst-1 gene among the study group, its distribution was similar in both MRSA and MSSA groups. Third, the more strains carrying the tsst-1 gene, further the disease susceptibility occurs. Moreover, the prevalence of resistant strains results in inefficient treatment and a higher rate of mortality.
The discrepancy between our findings and other records may be due to a difference in geographic regions. It has been revealed that the virulence gene profiles of S. aureus strains isolated from various locations are different. Since records are limited for the distribution of the mentioned genes in Iran and its neighbor countries, it is possible that the high frequency of strains harboring the tsst-1 gene and elevated drug resistance among them may be due to differences in geographic regions. Moreover, using a drug resistance pattern obtained from this work might be helpful for selecting a choice antibiotic in close areas. However, further investigations are needed.