1. Background
Staphylococcus aureus is one of the most common nosocomial pathogens which can cause a broad spectrum of infections, ranging from mild skin infections to severe abscesses, sepsis, endocarditis, osteomyelitis, urinary tract infections (UTI), and fatal necrotizing pneumonia (1). Staphylococcus aureus strains have the ability to become resistant to different classes of antimicrobial agents such as methicillin (2). Resistance to methicillin in S. aureus was first reported in 1961, just one year after its introduction, and methicillin-resistant Staphylococcus aureus (MRSA) has spread extensively worldwide during the last few decades (3). Resistance to methicillin is due to the presence of the staphylococcal cassette chromosome mec (SCCmec) element, which is composed of regulatory genes such as the mecA, C, I and R gene complex, and the ccr (cassette chromosome recombinase) gene complex, encoding the recombinase gene (4). Based on the presence of different regulatory and structural genes, 11 genetic classes of SCCmec have been recognized. The most important feature of MRSA isolates is their resistance to a broad spectrum of antimicrobial agents, which makes infections by these bacteria difficult to treat (5, 6).
Aminoglycosides are one of the classes of antibiotics that play an important role in the treatment of staphylococcal infections (7). The main mechanism of resistance to aminoglycosides is the inactivation of antibiotics by aminoglycoside-modifying enzymes (AMEs) that are encoded by genetic elements (7, 8). The aac (6’)-Ie + aph (2’’), ant (4’)-Ia, aph (3’)-IIIa, and ant (6)-Ia genes that encode aminoglycoside-6'-N-acetyltransferase/2"-O-phosphoryltransferase, aminoglycoside-4'-O-nucleotidyltransferase I, aminoglycoside-3'-O- phosphoryltransferase III, and streptomycin modifying enzyme, respectively, are hence the most important genes in this regard. Resistance to gentamicin, kanamycin, and tobramycin in staphylococci is mediated by a bi-functional enzyme displaying AAC (6') and APH (2") activity. The ANT (4')-IA enzyme inactivates neomycin, kanamycin, tobramycin, amikacin, and kanamycin, while the APH (3')-III, enzyme inactivates neomycin (8, 9).
Different genotyping and phenotyping methods are available for the typing of bacteria, and they are useful in studies of the stability and diversity of bacterial populations in investigations. The genotyping methods are highly discriminative, useful, and flexible, although they are very expensive and laborious, especially when comparing data obtained from many isolates (10). Phenotyping involves methods used in studying the biochemical characterization of bacteria. The Phene-Plate (PhP) system is an automated phenotyping method based on the quantitative measurement of the kinetics of biochemical reactions formed by bacterial metabolism in the presence of different substrates. Compared to genotyping methods such as PFGE and MLST, PhP typing is a very simple, rapid, and cheap method that provides the necessary information required for the subtyping of bacteria. However, for some cases in which PhP typing could not be performed, genotyping is required in order to obtain the necessary information (10, 11).
2. Objectives
This study aimed to describe the frequency and the antibiotic susceptibility patterns of clonal groups of gentamicin-resistant strains of MRSA isolated from a tertiary care hospital in Tehran, Iran.
3. Materials and Methods
3.1. Sample Collection and Identification of Bacteria
During January to November 2012, a total of 301 S. aureus strains were collected from a tertiary care hospital in Tehran, Iran. This hospital is ranked as one of the top hospitals in the country and it is located in the center of Tehran. All specimens were collected from hospitalized patients who showed infections 72 hours after admission to the hospital. Of the 301 strains, 139, 99, 23, 21, ten, and nine were isolated from wounds, urine, blood, sputum, cerebrospinal fluid (CSF), and eyes, respectively. All of the isolates were cultured on HiCrome aureus agar (Himedia, India) and then identified at the species level using species-specific nucA gene primers as described previously (12). The DNA of all the S. aureus strains was extracted using the boiling method as described previously (13). Moreover, a High Pure PCR Template Preparation kit (Roche, Germany) was employed for DNA extraction from the MRSA strains.
3.2. Antibiotic Susceptibility Testing
All of the 301 S. aureus strains were tested for susceptibility to oxacillin (1 µg) and cefoxitin (30 µg) (Mast Diagnostics, Merseyside, United Kingdom) on Muller-Hinton agar (Merck, Germany) according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (14). The susceptibility of the MRSA strains to 15 different antibiotics was determined. The antibiotics used were: kanamycin (30 μg), amikacin (30 μg), penicillin (5 μg), minocycline (30 μg), erythromycin (15 μg), clindamycin (2 μg), tobramycin (10 μg), rifampin (5 μg), sulfamethoxazole-trimethoprim (1.25 - 23.75 μg), linezolid (10 μg), quinupristin-dalfopristin (15 μg), ciprofloxacin (30 μg), neomycin (30 µg), gentamicin (10 μg), and tetracycline (30 μg). The minimum inhibitory concentrationsof oxacillin, gentamicin, and vancomycin were evaluated using a broth microdilution assay according to the guidelines of the CLSI (15).
3.3. Detection of Aminoglycoside-Resistant Genes
The presence of aac (6’)-Ie/aph (2”), ant (4’)-Ia, aph (3’)-IIIa, and ant (6)-Ia genes among the MRSA strains was examined using specific primers in a PCR assay as described previously (7, 16).
3.4. PhP Typing
All of the MRSA strains were typed using high resolution PhP-CS plates (PhPlate AB, Stockholm, Sweden). The biochemical fingerprinting method was performed as reported by Rahimi et al. (3).
4. Results
4.1. Identification and Antibiotic Resistance
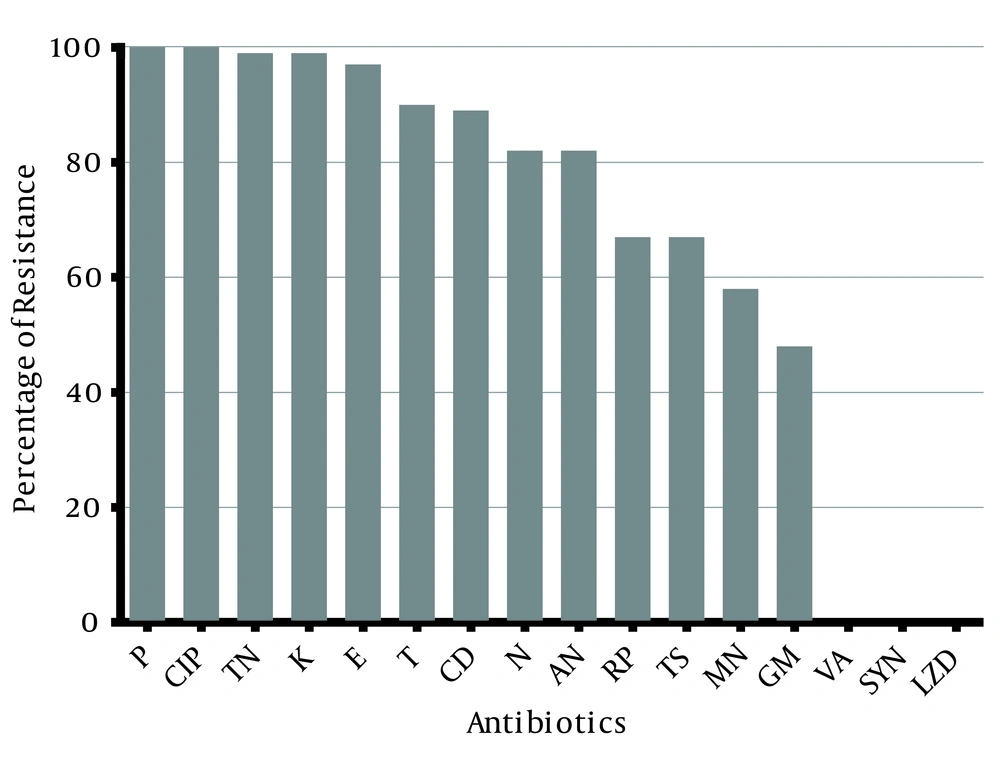
Of the 301 S. aureus strains isolated from the clinical samples, 90 strains (29.9%) were confirmed as MRSA using phenotypic and genotypic methods. These isolates originated from wounds (46%), urine (30%), sputum (13%), and blood (11%), respectively. The isolates were recovered from hospitalized patients and so were classified as hospital-acquired MRSA (HA-MRSA) strains. All of the isolates showed resistance to cefoxitin disc. The MRSA strains were resistant to penicillin (100%), ciprofloxacin (100%), tobramycin (99%), kanamycin (99%), erythromycin (97%), tetracycline (90%), clindamycin (89%), and amikacin (82%) (Figure 1). All of the strains were susceptible to vancomycin, linezolid, and quinupristin-dalfopristin.
Overall, 22 antibiotic resistance patterns were identified among the MRSA strains, of which 1.1% were resistant to at least three different antibiotics (Table 1). Of these, 73.3% of the isolates showed resistance to ten to 12 antibiotics. Moreover, six strains (6.7%) were resistant to all of the antibiotics tested except for vancomycin, linezolid, and quinupristin-dalfopristin.
| No. of Antibiotics | Values a | Pattern |
|---|---|---|
| Three antibiotics | 1 (1.1) | |
| P, CIP, TS | 1 (1.1) | 1 |
| Eight antibiotics | 4 (4.4) | |
| P, CIP, TN, K, E, CD, TS, RP | 4 (4.4) | 2 |
| Nine Antibiotics | 13 (14.4) | |
| P, CIP, TN, K, T, AN, N, RP, GM | 2 (2.2) | 3 |
| P, CIP, TN, K, E, CD, AN, N, TS | 2 (2.2) | 4 |
| P, CIP, TN, K, E, CD, AN, N, RP | 2 (2.2) | 5 |
| P, CIP, TN, K, E, T, TS, RP, MN | 1 (1.1) | 6 |
| P, CIP, TN, K, E, T, CD, TS, MN | 2 (2.2) | 7 |
| P, CIP, TN, K, E, T, CD, TS, RP | 2 (2.2) | 8 |
| P, CIP, TN, K, E, T, CD, RP, MN | 2 (2.2) | 9 |
| Ten antibiotics | 18 (20) | |
| P, CIP, TN, K, E, T, AN, N, TS, GM | 4 (4.4) | 10 |
| P, CIP, TN, K, E, T, CD, AN, N, RP | 8 (8.9) | 11 |
| P, CIP, TN, K, E, T, CD, TS, RP, MN | 4 (4.4) | 12 |
| P, CIP, TN, K, E, T, AN, TS, MN, GM | 2 (2.2) | 13 |
| Eleven antibiotics | 20 (22.2) | |
| P, CIP, TN, K, E, T, CD, AN, N, RP, GM | 2 (2.2) | 14 |
| P, CIP, TN, K, E, T, CD, AN, N, RP, MN | 13 (14.4) | 15 |
| P, CIP, TN, K, E, T, CD, AN, N, TS, GM | 3 (3.3) | 16 |
| P, CIP, TN, K, E, T, CD, AN, N, TS, RP | 2 (2.2) | 17 |
| Twelve antibiotics | 28 (31.1) | |
| P, CIP, TN, K, E, T, CD, AN, N, TS, RP, GM | 6 (6.7) | 18 |
| P, CIP, TN, K, E, T, CD, AN, N, TS, RP, MN | 4 (4.4) | 19 |
| P, CIP, TN, K, E, T, CD, AN, N, TS, MN, GM | 16 (17.8) | 20 |
| P, CIP, TN, K, E, T, CD, AN, N, RP, MN, GM | 2 (2.2) | 21 |
| Thirteen antibiotics | 6 (6.7) | |
| P, CIP, TN, K, E, T, CD, AN, N, TS, RP, MN, GM | 6 (6.7) | 22 |
aData are presented as No. (%).
The MIC range varied from 16 to 512 µg/mL, with 33% and 26% of strains having an MIC of 128 µg/mL and 256 µg/mL for oxacillin, respectively. Moreover, one strain had an MIC of 16 µg/mL. Also, for vancomycin, the MICs ranged from 0.06 to 0.5 µg/mL. Further, most of the strains (22.2%) had an MIC of 128 µg/mL for gentamicin, whilst 11 strains showed an MIC ≥ 16 µg/mL. Seven (7.8%), ten (11.1%), and 12 (13.3%) strains had an MIC of 1024, 512, and 256 µg/mL, respectively.
4.2. Detection of Aminoglycoside-Resistance Genes
All of the aminoglycoside-resistant strains were tested for the presence of different genes, with aac (6’)-Ie + aph (2’’) being found among 59 (65.6%) strains (Table 2) and hence being the most prevalent resistance.
| Pattern | Aminoglycoside-Resistant Genes | Phenotypes | Valuesa | |||
|---|---|---|---|---|---|---|
| Aac (6’)-Ie + aph (2’’) | ant (6)-Ia | ant (4’)-Ia | aph (3’)-IIIa | |||
| 1 | + | + | + | + | GM, K, AN, TN | 14 (15.6) |
| 2 | + | + | + | - | GM, K, AN, TN | 21 (23.3) |
| 3 | + | + | - | - | GM, K, AN, TN | 4 (4.4) |
| 4 | + | - | - | + | GM, K, AN, TN | 1 (1.1) |
| 5 | + | - | - | - | GM, K, AN, TN | 19 (21.1) |
| 6 | - | + | + | + | K, AN, TN, N | 1 (1.1) |
| 7 | - | + | + | - | K, AN, TN, N | 2 (2.2) |
| 8 | - | + | - | - | K, AN, TN, N | 1 (1.1) |
| 9 | - | - | - | + | K, AN, TN, N | 2 (2.2) |
| 10 | - | - | - | - | K, AN, TN, N | 25 (27.8) |
aData are presented as No. (%).
Forty-three (47.8%) isolates harbored the ant (6)-Ia resistance gene, while ant (4’)-Ia was detected in 38 (42.2%) strains. Moreover, the aph (3’)-IIIa gene was present in 18 (20%) strains. Also, 14 isolates (15.6%) carried four detected resistance genes together, and none of the genes were positive in 25 (27.8%) strains. On the other hand, aac (6’)-Ie + aph (2’’), aph (3’)-IIIa and ant (6)-Ia were detected alone in 19 (21.1%), two (2.2%) and one (1.1%) isolates, respectively.
4.3. PhP Typing
The PhP typing of the 90 MRSA strains revealed the presence of diverse (diversity index, DI = 0.818) PhP types among the isolates, consisting of seven common types (CT) and four single types (ST) (data not shown). Amongst the PhP types, CT 3 was the dominant type, with 31% of the isolates classified in this pattern. CTs 4 and 2 were also found among 24% and 11% of the isolates, respectively. Moreover, CTs 3 and 4 were common among all strains with different origins. On the other hand, the four STs were common among strains isolated from wounds and urine. The least number of strains belonged to CTs 5 - 7, which contained six isolates (7%).
5. Discussion
In this study, the frequency of MRSA in Tehran was 29.9%. Previous studies have revealed that the rate of MRSA in Iran varies from 19% to 90% in different cities (3, 13, 17-23). The variation seen in the different reports concerning Iran could be in part due to different populations, different geographical locations, and the quality of hospital sampling carried out. Of the 15 antibiotics tested in this study, all of the isolates showed susceptibility to vancomycin, linezolid, and quinupristin-dalfopristin, which is consistent with other studies in Iran (3, 13, 17-23). Although vancomycin is frequently used in a hospital setting, no vancomycin-intermediate S. aureus (VISA) or vancomycin-resistant S. aureus (VRSA) isolates were found in this study, which suggests that the increased use of certain antibiotics is not sufficient to ensure the appearance of resistant strains. Yet, most of the isolates were resistant to penicillin (100%), ciprofloxacin (100%), kanamycin (99%), tobramycin (99%), erythromycin (97%), tetracycline (90%), and clindamycin (89%), which indicates that these antibiotics are no longer effective antibiotics against MRSA infections in Tehran. In previous studies (3, 17, 19-23), a high rate of resistance to these antibiotics was reported. These antibiotics are used extensively in hospitals for the treatment of different infections and so the high rate of resistance is not surprising.
The rates of resistance tosulfamethoxazole-trimethoprim, rifampin, minocycline, and gentamycin in this study were also higher than those reported in other reports. This could be explained by the high level of these antibiotics being prescribed for the treatment of infections. Moreover, gentamicin is one of the most important antibiotics used in combination with other antibiotics worldwide for treatment of S. aureus infections (20, 24-26). In this study, similar to the findings of other reports (20, 24-27), the aac (6’)-Ie + aph (2’’) gene was dominant among the gentamicin-resistant strains of MRSA, and the isolates that were positive for this gene showed a high level resistance to gentamicin, which is consistent with other reports (16, 28-30). We also found 16 gentamicin-susceptible strains that harbored the aac (6’)-Ie + aph (2’’) gene. Although these isolates harbored the aac (6’)-Ie + aph (2’’) gene considered to be resistant to gentamicin and all aminoglycosides, we also found strains that were susceptible to gentamicin and showed resistance to other aminoglycosides, which is consistent with other reports (7). Hauschild et al. revealed that “detection of resistance genes in antibiotic susceptible strains is due to amplification of repressed antibiotic resistance gene or AME of these strains display lower enzymatic activity (7). Moreover, the prevalence rate of the ant (4’)-Ia gene was higher than in other reports (7, 16, 20, 26), which could be due to the high resistance to kanamycin, with 89 out of 90 strains being kanamycin-resistant. Also, we found a strain that was susceptible to all of the aminoglycoside antibiotics tested and was also not positive for all of the genes. Differences between reports from different countries could be due to differences among the isolates and different geographical regions. Our results illustrated that all of the aminoglycosides tested in this study are no longer effective agents against MRSA strains.
The results of the PhP typing showed the presence of diverse PhP types consisting of seven CTs and four STs, indicating that the presence of MRSA strains in this hospital in Tehran is attributable to the spread of a limited number of clonal types. CTs 3 and 4 were common between MRSA strains with different origins, which further supports the spread of these clonal types among strains collected from this hospital in Tehran. In another study in Tehran, more diverse PhP types (consisting of 18 CTs and 15 STs) were reported among the MRSA strains isolated from a tertiary care hospital (3). On the other hand, 16 CTs and 13 STs were identified in MRSA strains isolated from sewage treatment plants in Tehran (31). Javidnia et al. reported six CTs and 13 STs among MRSA strains in Tehran (19). Seven CTs identified in this study were also common among the MRSA isolates reported in other studies, in which CT 3 was the predominant type and its dissemination is consistent with previous studies in Tehran (3, 9, 31, 32). In this study, different PhP types were common among different samples, which indicates the prevalence of MRSA strains in different wards of the hospital and also highlights the presence of common CTs in this hospital. The MRSA strains isolated from wounds and urine were grouped in different CTs and showed the highest frequency. A certain PhP MRSA clonal type, i.e. CT 3 (comprising 31% of the isolates), was identified in various samples tested, indicating an epidemiological link between the MRSA strains isolated from different sources. The results indicate that MRSA strains are endemic in this hospital in Tehran; therefore, contamination of patients during the hospitalization process may be responsible for these infections.
As previously reported, the aminoglycoside-resistant HA-MRSA strains have spread widely through the community and identical HA-MRSA strains from different sources in Iran have been isolated, indicating the hospital origin of these strains. In conclusion, our results showed that the PhP typing method provided useful information for clonal dissemination and also for the epidemiological links of clonal groups of aminoglycoside-resistant strains of MRSA in hospitals in Tehran. Moreover, the presence of predominant clonal types among all strains isolated from different sources with different antimicrobial resistance patterns suggested that cross-infection was a factor in the maintenance of these strains in hospitals in Tehran.