1. Background
Crimean-Congo hemorrhagic fever (CCHF) belongs to the Nairovirus genus. This agent causes severe disease in humans (tick borne zoonotic disease), and is endemicto many areas of Africa, Asia, and Europe (1-6). For example, this agent has caused serious challenges in the Sistan-va-Baluchestan province of Iran (7-15). The mortality rate is about 40%, but can vary from 10% to 50% (16, 17). Since there is no approved vaccine or specific treatment for CCHF, an early and accurate diagnosis, as well as reliable surveillance are essential for patient improvement, case management, and the protection of the medical staff diagnostic assays for the CCHF virus include viral isolation, the enzyme linked immunosorbent assay (ELISA), and reverse-transcription polymerase chain reaction (RT-PCR) (4, 18-21). Viral RNA from different clinical samples is an appropriate detection target during the acute phase of an infection, or even before the onset of illness, when antibody detection is unreliable or impossible (22, 23). Unlike the traditional RT-PCR method, the real-time RT-PCR (rRT-PCR) does not require post-PCR sample handling, preventing the PCR product dependent eventual contamination transmission, resulting in much faster and higher throughput assays. It is extremely accurate, more sensitive, and less labor-intensive than the current, traditional RT-PCR methods, and has therefore become a accepted diagnostic test for the recognition of many microorganisms (24). In this study, we developed a novel, in-house, SYBR Green based one-step rRT-PCR assay for the rapid and accurate diagnosis of CCHF in suspected patients from the Sistan-va-Baluchestan province of Iran.
2. Objectives
The aim of this research was to evaluate the application of a novel SYBR Green based one-step rRT-PCR assay, developed for the diagnosis of the CCHF virus.
3. Patients and Methods
3.1. Sample Collection
In order to accomplish this research, we prepared 75 cell-free serum samples from different CCHF suspected patients from the Sistan-va-Baluchestan province in southeast Iran. Most of these samples were already confirmed positive for CCHF viral RNA via RT-PCR analysis (Pasteur Institute, Tehran, Iran), and they had been stored at -80°C.
3.2. Viral RNA Extraction
Viral RNA was extracted from 200 µl of each serum sample by using a High Pure Viral RNA Kit (Roche-Germany) within 30 minutes, according to the manufacturer's instructions. Then, the extracted RNA was dissolved in 50 µl of RNase-free water and stored at -80°C until analysis (only 5 µl was used in each assay). It is notable that all of the manipulations were carried out in a cabinet with class II biosafety.
3.3. Design and Synthesis of Specific Primers
The highly conserved S-region sequence of the CCHF viral genome was adapted from GenBank, after the alignment of the nucleotide sequences from the available CCHF viral strains(obtained from the NCBI database). Then, the specific primers targeting this region were designed using the Allele ID software, version 7.0 (Premier Biosoft International, Palo Alto, CA, USA). The forward oligonucleotide primer for the S-region internal sequence was 5'-GAA ACA ACG GAG GTG GTC TCG-3', and the reverse oligonucleotide primer for the S-region internal sequence was 5'-ACC ACG TCC CTC AAA CAG-3'. The region targeted by these primerswas from nucleotide 251 to 670 for the S-sequence, as one of the conserved regions of the viral genome. Finally, the primers were synthesized by the Bioneer Company (Republic of Korea).
3.4. One-Step Real-Time RT-PCR
The novel, in-house SYBR Green based one-step rRT-PCR assay was developed using the QuantiFast SYBR Green RT-PCR Kit (Qiagen, Hilden, Germany), using the Corbet (Rotor-Gene) 6000 (Qiagen, Germany) and Applied Biosystems (ABI) 7500 (USA) real-time PCR instruments. The amplifications were accomplished within 1 hour , in 20 µL reaction mixtures containing 5 µL of the targeted viral RNA and 2 µL (20 pmol) of each primer (10 µM). The cycling conditions were as follows: a single cycle of 10 minutes at 50°C and 3 minutes at 95°C, followed by 40 cycles of 10 seconds at 95°C, and a final cycle of 30 seconds at 60°C (an analysis of fluorescence was performed at the end of each 60°C step). The rRT-PCR products were detected via an increase in fluorescence from cycle to cycle, and the positive and negative controls were used as the quality controls for the process. Both the cycling and melting curves in the real-time analysis were evaluated with respect to the negative and positive controls. The rRT-PCR amplicons were then confirmed via electrophoresis.
3.5. Evaluation of the Analytical Sensitivity and Specificity of the Assay
3.5.1. Analytical Sensitivity
In order to determinate the limit of detection (LoD), serial dilutions containing 2, 20, 200, and 2000 copies of the positive control sample per reaction were analyzed us in real-time PCR. Each assay was evaluated by at least three different users, three times each, on different days. The highest dilution of sample at which all of the replicates were positive was defined as the limit of detection (LoD) for this assay. The LoD of the assay signature was 20 or fewer copies of the viral genome/reaction. The serum samples of the different CCHF patients confirmed to be positive by the Pasteur Institute (Tehran, Iran) were used as positive control samples, and no false-positives were detected in the sequencing results. In fact, of these positive samples confirmed by sequencing, some were selected as positive control samples.
3.5.2. Analytical Specificity Viain Silico Prediction
As mentioned in section 3.3, none of the primer sequences showed genomic cross-reactivity with other viruses, the human genome, or other probable interfering genomes in a BLAST search analysis, and only a 420 bp fragment of the CCHF viral S-region was detected (results not presented).
3.6. Repeatability of Assay
All of the evaluations were performed with assay replications by at least three different users, three times each, on different days, and the repeatability of the assays was confirmed. In addition, the assays were developed on the Corbet (Rotor-Gene) 6000 (Qiagen, Germany) and Applied Biosystems (ABI) 7500 (USA) real-time PCR instruments.
4. Results
4.1. Diagnostic Test of Viral RNA in Suspected Serum Samples
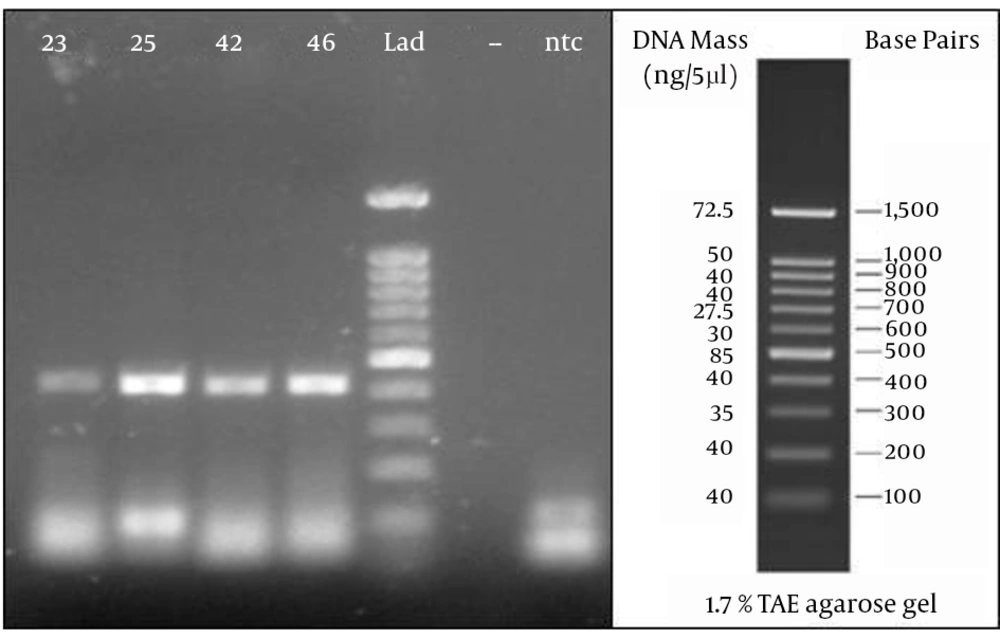
From a total of 75 suspected serum samples, 42 were confirmed to be positive and 33 were negative for the CCHF virus using this novel real-time RT-PCR, and no false-positives were detected in the sequencing results. In all of the positive samples, the predicted length of 420 bp was confirmed by electrophoresis (Figure 1).
4.2. Melting Curve Analysis
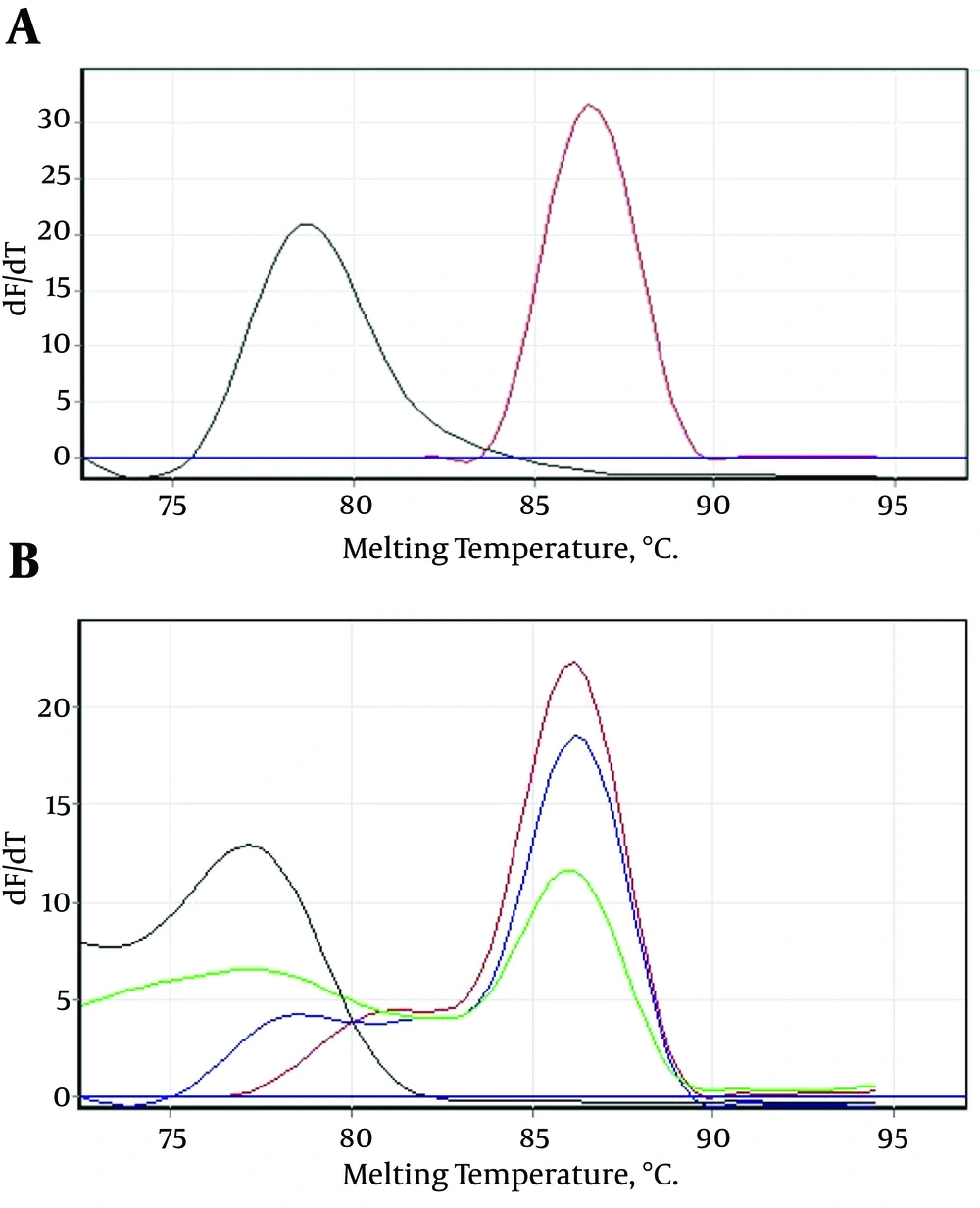
After 40 amplification cycles, the melting curve analysis revealed a mean melting temperature (Tm) of 86.5 ± 0.6°C (quite different from that of the primer-dimers) (Figure 2A and B), and the positive samples showed only a small variation in the parameters.
A, Showing that the melting temperature (Tm) of the specific amplicons was 86.5°C, and different from the primer-dimers; B, serial dilutions of the positive control sample showing that the mean melting temperature (Tm) of the specific amplicons was 86.5 ± 0.6°C, and the melting temperature (Tm) of the primer-dimers was different.
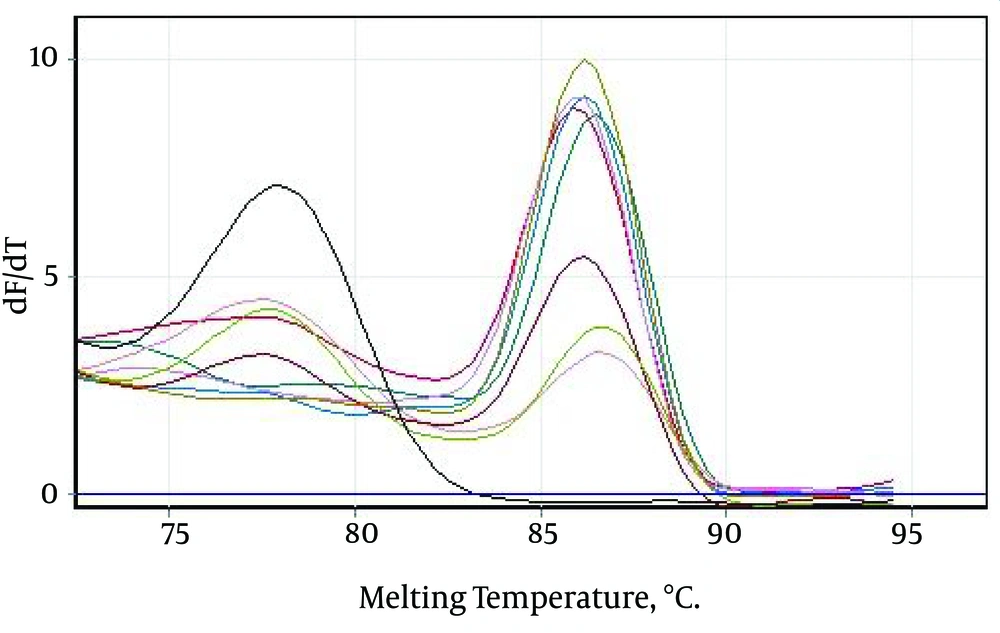
Only those Tm peaks between 86.5 ± 0.6°C were accepted as positive (observed in most of the tested serum samples), as shown in the melting curves in Figure 2A and B. The positive samples showed only a small variation in the parameters (Figure 3).
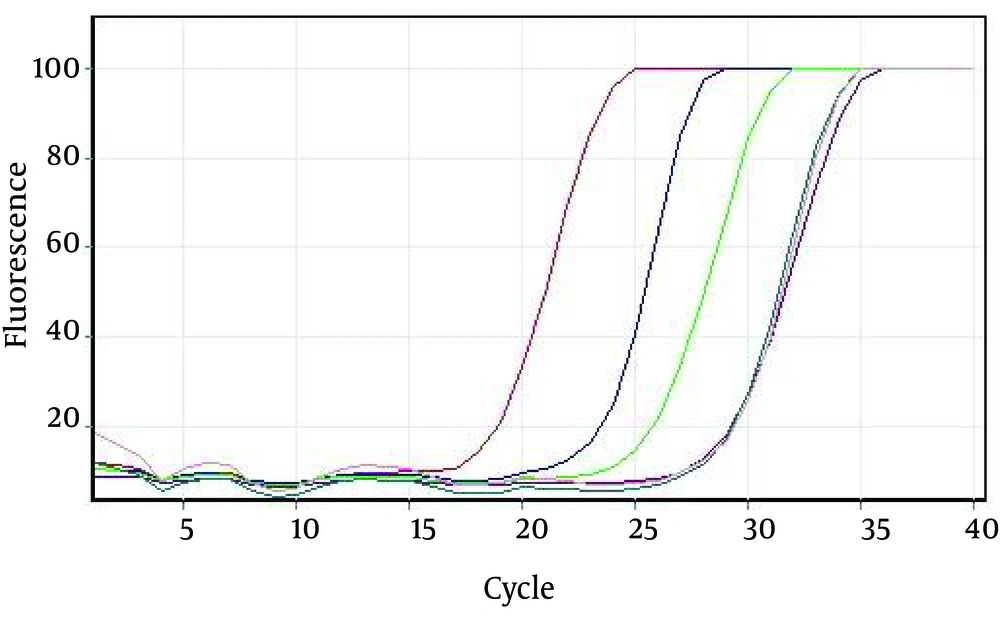
4.3. Evaluation of Analytical Sensitivity of Assays
We prepared serial dilutions of the positive control sample for the evaluation of the analytical sensitivity of the assays (Figure 4). The serial dilutions were analyzed using rRT-PCR.The LoD of the assay signature was 20 or fewer copies of the viral genome/reaction (Table 1)
| Serial Dilution (Copies/Reaction) | Positive Tests (%) S-Region |
|---|---|
| 2 | 34 |
| 20a | 100 |
| 200 | 100 |
| 2000 | 100 |
CCHF Viral Rrt-Pcr Assay Limit of Detection with Positive Control Sample.
5. Discussion
CCHF is an acute, tick-borne zoonotic disease, which is characterized by a fulminant and often hemorrhagic course of illness, with a case fatality rate of 10 - 50% (16, 17) Viral RNA from different clinical samples is an appropriate detection target during the acute phase of an infection, or even before the onset of illness, when antibody detection is unreliable or impossible (22, 23). Since there is no an approved vaccine or a specific treatment for CCHF viral infections, an early and accurate diagnosis is essential for patient improvement and case management. Various detection methods, targeting viral genomes, have been developed for the rapid identification of different viral diseases, but RT-PCR has become the most popular molecular diagnostic approach in clinical laboratories due to its simplicity, high sensitivity, and high specificity (23). Traditional RT-PCR methods require amplification in a thermo cycler and product analysis via gel electrophoresis, both of which are time-consuming and laborious (25).
By eliminating the need for post-amplification product processing, real-time RT-PCR enables a shortened turnaround time for reporting results, which is critical for deciding on the isolation of suspected cases and contact tracing. Real-time RT-PCR assays are sensitive and rapid, and can be automated. They also have greater reproducibility when the need for post-PCR processing is eliminated, thus preventing carry-over contamination (25). In this novel, in-house SYBR Green based one-step rRT-PCR assay, the products were identified based on a Tm curve analysis, and the PCR products from each primer pair were generated based on the individual Tm values. After 40 amplification cycles, the melting curve analysis revealed the mean melting temperatures (Tm) of 86.5 ± 0.6°C (quite different from those of the primer-dimers), while the positive samples showed only a small variation in the parameters. The sensitivity test showed that this assay can detect less than 20 copies of viral RNA per reaction. The SYBR Green assay not only provides confidence in the identification of the target genes, but also reduces the risk of laboratory product contamination. This is because the amplification reaction, detection of the PCR products, and the Tm analysis can be performed in a single tube (26). The SYBR green based RT-PCR assay is less influenced by mismatches, due to probe independency, and has the advantages of relative simplicity and lower running costs, which may be more appropriate for use in first line clinical laboratories (23).
5.1. Conclusion
This study has shown that this newly developed one-step rRT-PCR assay is a rapid, reliable, repeatable, specific, sensitive, and simple tool for the detection of theCCHFvirus.