1. Background
Helicobacter pylori is Gram negative, curved microaerophilic bacilli and a major cause of gastric ulcer, duodenal ulcer, gastritis, Mucosa-associated lymphoid tissue (MALT) lymphoma and associated with gastric adenocarcinoma (1-3). The prevalence of H.pylori varies among different countries, for example, in many developing countries such as Iran, it is more than 70%, whereas in developed countries it is up to 50% (4). Antibiotic treatment of H. pylori infection is complicated, requiring multiple antibiotics along with acid inhibitors to ensure high cure rate (5).
Clarithromycin is one of the key antibiotics for the treatment of H.pylori. This macrolide antibiotic has been used frequently in combination with other antimicrobial agents for the treatment of H. pylori infection (6). Unfortunately, resistance to this antibiotic is increasing worldwide and failure treatment is attributed to acquired antibiotic resistance of the organism or lack of compliance with the antibiotic regimen (7). The prevalence of H. pylori resistance to clarithromycin varies among different countries; 12% in Japan, 1.7% - 23.4% in Europe and 10.6% - 25% in north America (5, 6, 8). Antimicrobial activity of clarithromycin is attributed to inhibition of protein synthesis via attaching to bacterial ribosomal 50S subunit (9). It seems the clarithromycin resistance in H.pylori is associated with point mutations in the 23SrRNA and inhibition of protein synthesis by detachment of peptidyl-tRNA from the ribosome during elongation (10). Adenine to guanine transitions at positions 2142 and 2143 are the main 23SrRNA mutations in clarithromycin-resistant isolates (6, 11). All these mutations have been shown to confer resistance to this macrolide by mutagenesis study (12). Other mutations that have been observed in clarithromycin-resistant H. pylori isolates are A2515G and T2717C, A2116G, G2141A, A2144T, T2182C, G2224A, C2245T and T2289C (13). The A2142C/G and d A2143G mutations also generate restriction sites for MboII and BsaI restriction endonuclease, which may be used for rapid screening of clarithromycin resistance (14).
Since susceptibility tests and culture of H.pylori is difficult and time consuming, rapid molecular detection tests such as fluorescent in situ hybridization, reverse hybridization line probe assay, sequencing, real-time polymerase chain reaction (PCR) and EIA of DNA have been developed (15). In the past decade different PCR-based methods have been developed as alternative tools (7). The aim of this study was to determine A2142G and A2143G mutations by PCR-RFLP in H.pylori isolates from patients with gastric diseases in Rasht, North of Iran.
2. Objectives
The aim of this study was to determine the frequency of A2143G point mutation in 23SrRNA of H. pylori strains isolated from gastric biopsies of patients in Rasht, north of Iran, by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
3. Patients and Methods
3.1. Patients
Between April 2012 and February 2013, gastric biopsy specimens were taken from patients with gastric disorders, who had an indication for endoscopy before receiving anti H. pylori treatment. Informed consent was obtained from all patients. One specimen was taken for a rapid urease test (RUT) and another in normal saline for culture.
3.2. Culture of Helicobacter pylori and E-test
Biopsies were cultured on Columbia agar medium supplemented with 10% Fetal calf serum (FCS), 5% sheep blood and 10 g/mL trimethoprim, 6 g/mL cefsulodin and 5 g/mL vancomycin and incubated under microaerophilic condition at 37°C for five to seven days. After observing colonies, Gram negative curved bacilli with catalase, oxidase and urease positive tests were confirmed as H.pylori (16). Suspensions from primary isolates were prepared in sterile normal saline equal to standard 4 McFarland (12 × 108 CFU/mL). Muller–Hinton agar plates with 5% sheep blood without any antibiotics were streaked in three directions with a sterile swab dipped into each bacterial suspension to produce a lawn of growth. E-test strips (BioMerieux, SA, France) were placed onto separate plates, which were immediately incubated under microaerophilic condition at 37°C for 72 hours. Isolated strains were tested for clarithromycin resistance using a breakpoint at minimum inhibitory concentration (MIC) of ≥ 1 mg/L (3). Finally, isolates were harvested in Brucella broth containing 20% glycerol and stored at -80°C.
3.3. Identification of Helicobacter pylori by the Polymerase Chain Reaction
DNA was extracted from fresh isolates of H.pylori before storage at -80°C using a DNA extraction kit (Roche Co., Germany), according to the manufacturer’s instructions. Then, DNA concentration was assessed by Nanodrop. All extracted DNA were amplified for ureC (glmM) gene by PCR, using an automatic thermo cycler (Eppendorf Personal 5332, Germany).
Polymerase chain reactions were performed in a final volume of 25 μL containing 2.5 μL 10x buffer, 0.75 μL MgCl2 (50 mM), 2.5 unit Taq DNA polymerase (Cinnagene CO., Iran), 0.5 μL dNTP (10 mM) (Cinnagene Co., Iran), 20 Pico mole of each primer and 1 μL of genomic DNA as a template. These primers (Table 1) were used to amplify a fragment of 294 bp from the ureC gene (17). Amplification was performed with pre-incubation (94° for three minutes), followed by 35 cycles of denaturation (94° for one minute), annealing (55°C for one minute), extension (72°C for one minute) and a final extension (72°C for five minutes). The PCR products were electrophoresed in 1.5% agarose gel (Roche, Germany) containing Sybrsafe. A DNA ladder (Roche Co, Germany) was used to detect the molecular weights of observed bands under a UV lamp. Results were sent to the Bioneer Company in South Korea for sequencing to confirm whether the observed bands were related to the mentioned genes.
3.4. Amplification of 23SrRNA and Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP)
Extracted DNA from H.pylori isolates were used as templates for amplifying a 429 bp fragment of peptidyl transferase of 23SrRNA gene (GenBank accession number U27270), using specific primers (Table 1). The PCR reaction was performed in a final volume of 25 μL containing 2.5 μL 10x buffer, 0.75 μL MgCl2 (50 mM), 2 unit Taq DNA polymerase (Cinnagene CO., Iran), 0.5 μL dNTP (10mM) (Cinnagene Co., Iran), 10 Pico mole of each primer and 1 μL of genomic DNA as a template. Amplification was performed with pre-incubation (95°C for five minutes), followed by 35 cycles of denaturation (95ºC for 30 seconds), annealing (54°C for 30 seconds), extension (72°C for 30 seconds) and a final extension (72°C for 10 minutes). The PCR products were electrophoresed as mentioned previously.
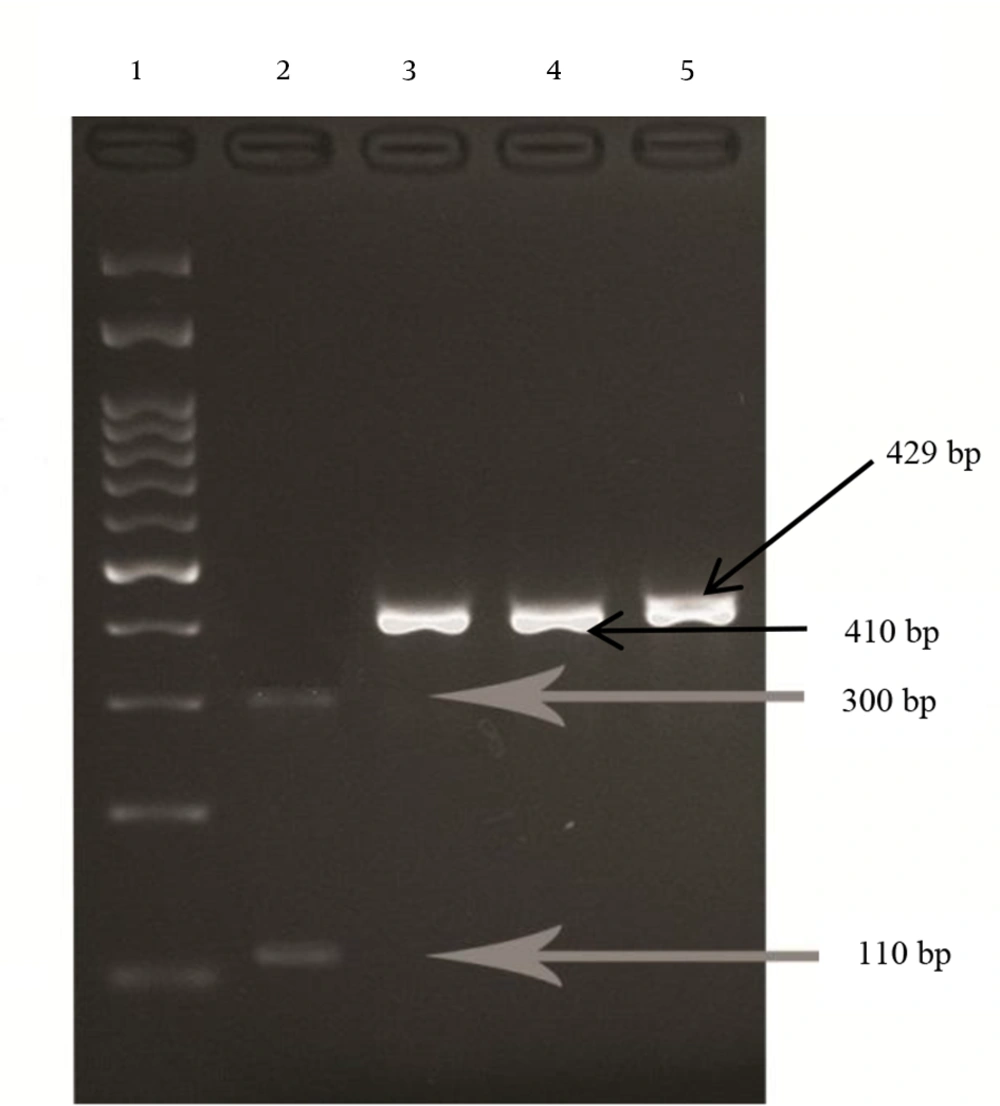
To detect the A-G point mutations at positions 2142 and 2143, the PCR products were digested with the MboII and BsaI (MBI Fermentas, Lithuania) restriction enzymes, respectively, according to the manufacturer’s instructions, and analyzed on 2% agarose gel containing Sybrsafe. The 429 bp fragment normally has one restriction site for BsaI. If the gene is wild type, BsaI finds one digestion site and produces two fragments (19 and 410 bp), but if the A2143G mutation has occurred in the 429 bp fragment, the BsaI finds two restriction sites and produces three fragments (19, 110 and 300 bp). The 429 bp fragment normally has no restriction site for the MboII enzyme. Therefore, if the gene is wild type, the 429 bp fragment remains undigested, but if the A2142G mutation occurs, MboII finds one restriction site and produces two fragments (336 and 93 bp). The results were confirmed by sequencing of PCR products 23SrRNA gene in wild type and mutant strains via the Bioneer company.
3.5. Statistics
Normal distribution was assessed the by Kolmogorov-Smirnov test. Prevalence rate and qualitative variables were assessed by percentages, and quantitative variables were reported as mean ± standard deviation (SD). The Fisher exact test and X2 were applied for qualitative variables and one-way analysis of variance (ANOVA) for quantitative variables. The SPSS version16 was used for analyzing the data. The P < 0.05 was considered significant.
4. Results
Among the 89 clinical isolates of H. pylori (43 isolates from females and 46 from males, mean age 53.65 ± 17.89), 30 strains (33.7%) were from gastritis samples, seven (7.9%) from gastric ulcer, 24 (27%) from duodenal ulcer, 15 (16.8%) from non-ulcer dyspepsia (NUD) and 13 (14.6%) from gastric adenocarcinoma. A 294 bp fragment was amplified in all H.pylori isolates after amplification of the ureC (glmM) gene.
According to the chi square test, there was no significant correlation between different gastric disorders and gender (P = 0.986 and X2 = 0.352). One way analysis of variance (ANOVA) test revealed a significant difference between mean age of patients infected with H.pylori and different gastric disorders (P = 0.0001). Among 89 isolates, five (5.6%) isolates were resistant to clarithromycin. The BsaI restriction enzyme cut the PCR products of the 23SrRNA gene in five (5.6%) resistant strains to 19 bp, 110 bp and 300 bp bands (Figure 1), indicating that the five resistant strains had an A to G mutation at position 2143 (A2143G). In susceptible strains (wild type) the enzyme had only one restriction site and cut the PCR product to two fragments of 19 bp and 410 bp. None of the 89 isolates of H.pylori were cut with the MboII enzyme, which means there was no A to G mutation at position 2142. The highest resistance rate was found in females (7%) and the most sensitivity rate was found in males (95.7%). The Fisher exact test revealed that there was no significant relationship between clarithromycin resistance and gender (P = 0.67).
Lane 1, DNA ladder (100 bp); Lane 2, clarithromycin resistant strain digested with BsaI at two sites; Lane 3 and 4, wild types of H. pylori (susceptible to clarithromycin) digested with BsaI, with one digestion site; Lane 5, wild type (susceptible to clarithromycin) treated with MboII with no digestion site.
The highest sensitivity rate was seen in H.pylori isolated from NUD and gastric adenocarcinoma (100%) and the highest resistance rate was seen in H.pylori isolated from gastric ulcer (14.3%). There was no significant relationship between resistant strains and different types of gastric disorders based on the chi-square test (P = 0.549 and X2 = 3.5). The PCR-RFLP results agreed with the antibiogram and E-test results.
5. Discussion
The increased use of antibiotics to treat other diseases such as parasitic, respiratory, intestinal infections and etc. has caused more failures in curing H.pylori infections. Usually, the increased resistance level is consistent with antibiotic consumption pattern (18). Among the antibiotics used to treatment H.pylori infection, clarithromycin has the most powerful bacteriostatic effect. Clarithromycin is a member of the macrolide family, which acts through connection to peptidyl transferase of 23SrRNA and prevents bacterial protein synthesis. An ignorance in treatment or lack of followed up will lead to resistance to this antibiotic and under some conditions may have some severe side effects such as gastric adenocarcinoma (2, 7). It is very important to understand the molecular mechanism of resistance, because it can facilitate the development of molecular techniques. Polymerase chain reaction-RFLP is one of these techniques, which was used in this study to illustrate that only the strains with A2143G mutation are digested when exposed to BsaI. The level of resistance to clarithromycin varies from country to country even from region to region in a single country. For instance, the prevalence of resistance to clarithromycin in Iran (Jahrom, Kerman and Tehran) was 23.78%, 31.7% and 21.7%, respectively (19, 20), which compared to our study (5.6%) had a higher prevalence. In the present study, the resistance prevalence among the isolated strains was low. This was the first study ever conducted in the Guilan province (north of Iran). There are some point mutations in association with the resistance to clarithromycin. The most common ones are A2142G and A2143G, and according to our research as well as some studies, the most abundant mutation was A2143G (2, 3, 11, 21).
In this study, resistance to clarithromycin from the individual’s gender aspect was also examined. Although the relative abundance of resistance in females was higher than males yet based on the investigated individual’s gender, there was no significant correlation with clarithromycin resistance due to mutation at A2143G. This is consistent with the findings of Alarcon and others (2, 7, 20, 21). In the study of Abdollahi et al. there was a significant relationship between the gender of the patients and the mutation of A2143G, so that the above-mentioned mutation was only observed in females (22). These findings were not consistent with our results.
In this study we also assessed the rate of relative abundance of resistance to clarithromycin due to A2143G mutation, according to the patients’ age. Although age is proportional with resistance yet based on the individual’s age, a significant correlation between the resistance status due to A2143G mutation was not observed. This coincides with Raymond et al. and Abdollahi et al. studies (2, 22). Alarcon et al. believed that as patients’ age increased the level of resistance decreased (21). This finding does not agree with ours. In this study beside the relationship between age and gender, we also considered the correlation between various diseases and resistance to clarithromycin. It is true that there was no significant relationship between various diseases and the status of clarithromycin resistance, which is consistent with the findings of De Francesco et al. and Abadi et al. (7, 20), yet the highest relative abundance of resistance to clarithromycin was seen in the patients with gastric ulcer. This is in agreement with Abadi et al. and colleagues (20). In comparison to Abadi et al. and De Francesco et al., we assessed more diseases in our study; gastritis, gastric ulcer, duodenal ulcer, non-ulcer dyspepsia and gastric adenocarcinoma.
The difference between our results and the results of other studies may be due to the kind of diseases present in patients, from which samples were obtained.
Also, the lack of A2142G mutation in our study somehow can explain why resistance to clarithromycin is low in this province. Since the number of patients examined in this study was not large enough, the results of this study should be cautiously analyzed when compared with others.
We concluded that clarithromycin resistance in H.pylori strains isolated from the Guilan province is low and there was no mutation among the sensitive strains.