1. Background
Over the several last decades, nontuberculous mycobacteria (NTM) have been reported as some of the most important causes of pulmonary and non-pulmonary infections, which are explained in part by an increased number of susceptible immunocompromised individuals, such as those suffering from AIDS (1). The criteria for the treatment of NTM infections are based on the species involved, the immune characteristics of the patient, and the clinical manifestation of the infection (2). Nontuberculous mycobacteria are often resistant to first-line anti-tuberculosis drugs, which emphasizes the importance of species identification in mycobacterial diseases. Nontuberculous mycobacteria infections with slow-growing species are often treated by a three-component or dual therapy involving oral clarithromycin, rifabutin, ciprofloxacin, rifampicin, and ethambutol (3).
Rapidly growing mycobacteria are usually sensitive to drugs such as new-generation macrolides, cephalosporin, and some fluoroquinolones. Recent studies have indicated that the newest class of antibiotics, the fluorinated quinolones, have a good reaction against rapidly growing non-mycobacteria (4, 5). However, when a mycobacterial infection is suspected, quick and exact species-level identification of the mycobacteria is the most important step toward successful disease therapy. Ciprofloxacin acts by inhibiting DNA gyrase, an enzyme essential to separating the DNA of the bacteria (6). Characterization of the gyrA gene indicates that ciprofloxacin, in particular, acts as a barrier of cell division. Mutations in the gyrA site confer conformational shifts to imperfect binding of the medicine and finally results in resistance.
Many methods have been used to find these mutations. The gyrA locus from mycobacterial spontaneous sequencing has been clearly applied to characterize mutations related to resistance, but a number of other methods, such as PCR single-strand conformational polymorphism (PCR-SSCP), have also been successfully used (7). PCR-SSCP analysis implicates amplification of a part of the gene encoding for the specific object, and evaluation of the PCR products of medicine-sensitive and medicine-resistant strains by non-denaturing electrophoresis, in which mutations generally occur in a changed pattern (8, 9). In several parts of the world, different types of mutations have been identified in ciprofloxacin-resistant mycobacterial isolates. Most of these were point mutations involving the 83 codon (2).
2. Objectives
In this study, we determined the mycobacterial species with multiplex PCR and used PCR-SSCP and a direct sequencing analysis for detecting mutations in the DNA of the gyrA gene of ciprofloxacin resistance in clinical and environmental isolates of nontuberculous mycobacteria in the geographical region of Isfahan province in Iran. Based on a search of all of the available resources and databases in Iran, we could not find any studies similar to ours.
3. Materials and Methods
3.1. Mycobacterial Species
A total of 41 environmental and clinical isolates of NTM were collected from the microbial collection of the Isfahan University of Medical Sciences microbiology department and Isfahan tuberculosis. Isolates were separated from water, blood, abscess, and bronchial samples. There were 21 environmental isolates and 20 clinical isolates.
3.2. Primary Identification of Isolates
All isolates were subcultured on lowenstein-jensen medium (Merck/Germany) at 37°C for four weeks, and were then characterized by conventional methods, including colony morphology, pigmentation, growth temperature, speed of growth, ziehl-neelsen staining, and the nitrate-reduction test, as reported previously (10).
3.3. Identification of Isolates by Multiplex PCR
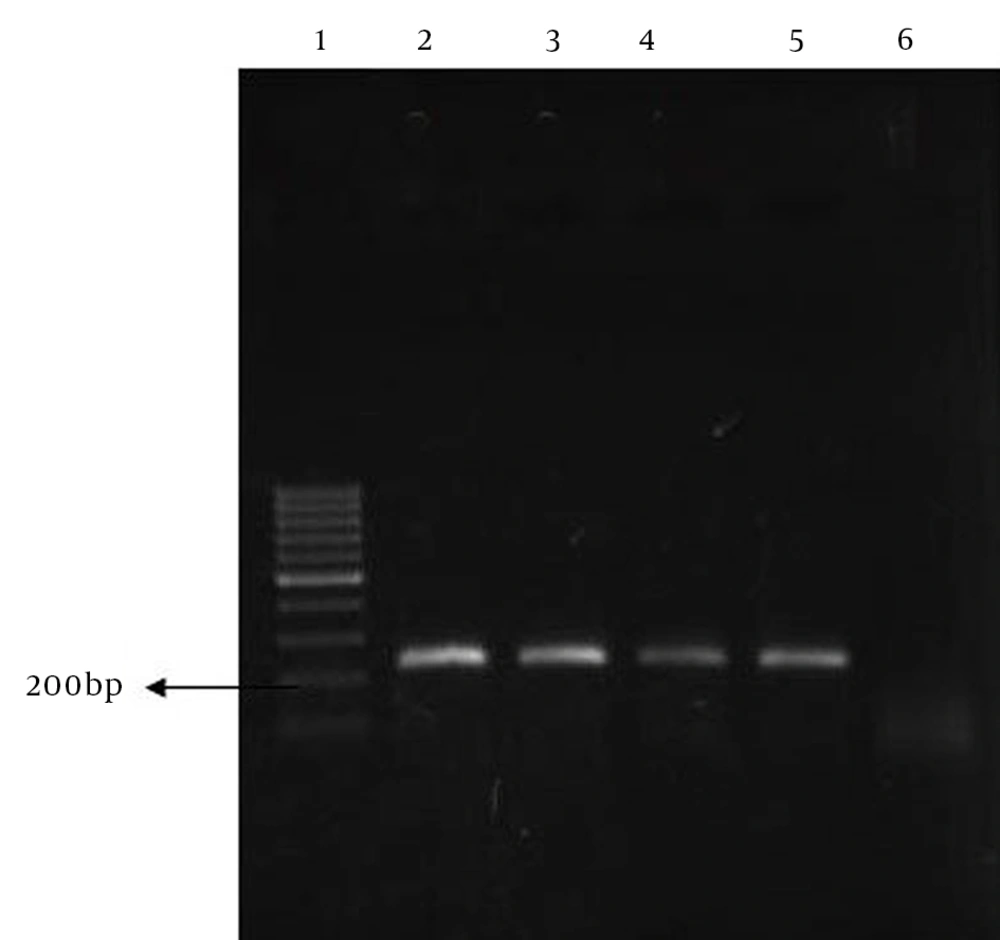
The chromosomal DNA of the isolates was extracted with the DNA mini-prep procedure, using hexadecyltrimethylammonium bromide (CTAB) as previously described, and a high pure PCR template preparation kit (Roch Applied Science, Germany). Concentration and purity of the extracted DNA was determined using a UV-photometer (Biometra, Germany) at 260 nm and 280 nm. Purified DNA was stored at -70°C until the PCR experiments, and about 10 ng of the extracted DNA was used for PCR amplification (Eppendorf, Germany). A multiplex PCR based on different housekeeping genes, using seven primers, was used for detection of Mycobacterium species. Genetic targets for the primer design included the 16S rDNA gene of all members of the genus Mycobacterium: the internal transcribed spacers of the Mycobacterium tuberculosiscomplex (MTC) (Mycobacterium tuberculosis, M. bovis, M. bovis BCG, and M. africanum), M. fortuitum, M. avium complex, and M. kansasii; the dnaJ gene of M. abscessus; and the gyrB of M. gordonae, which are deposited in the NCBI, DDBJ, and EMBL databases. The amplification reaction consisted of the following steps: one denaturation cycle at 95°C for 5 minutes, then 40 cycles of amplification at 95°C for 10 minutes, 60°C for 40 seconds, and 72°C for 1 minute, followed by one elongation cycle at 72°C for 10 minutes. The size of amplicons was then analyzed by UV detection of ethidium bromide agarose gel (11).
3.4. Susceptibility Testing
Colonies from all of the isolates were gathered from the lowenstein-jensen media and suspended in PBS, then thoroughly homogenized by shaking in glass bottles. Subsequently, a McFarland 1.0 concentration of a suspension of each isolate was prepared. For each isolate, 100 µL of 10-2 and 10-4 diluted suspension was inoculated on 7H10 agar medium enhanced with 10% oleic acid-albumin-dextrose complex (OADC). Serial three-fold concentrations of ciprofloxacin in media (1 µg/mL, 2 µg/mL and 4 µg/mL) and an antibiotic-free medium as the control were prepared and inoculated by diluted suspension of mycobacterial isolates. After seven days of incubation (for rapidly growing mycobacteria) and/or 14 - 21 days (for slow-growing mycobacteria) at 37°C, bacterial growth was measured. The lowest concentration of the drug that inhibited more than 99% of the mycobacteria was considered to be the minimum inhibitory concentration (MIC) (12).
3.5. PCR-SSCP Analysis of the Quinolone Resistance-Determining Region of the gyrA Gene
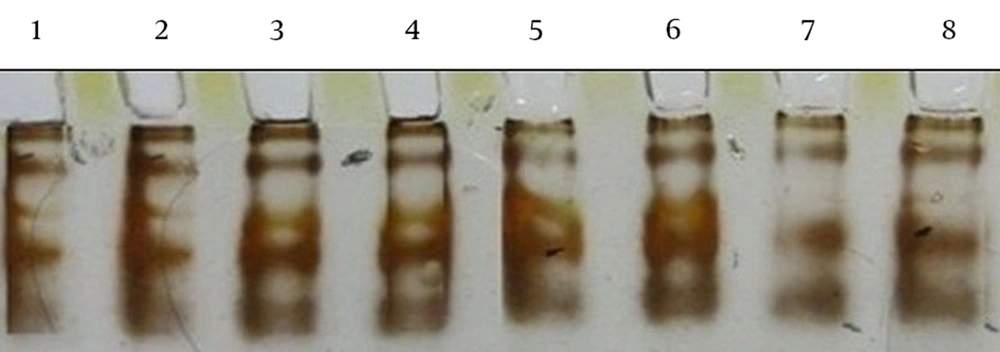
Extraction of mycobacterial DNA was performed as previously described. Amplification of the mycobacterial gyrA gene was done using primers F (5-CGCCGCGTGCTG/CATGCA/GATG-3) and R (5-C/TGGTGGA/GTCA/GTTA/GCCC/TGGCGA-3) (Bioneer/Korea) (13-15). PCR was performed in a 50 µL reaction mixture containing 1.0 µL of each primer (10 pmoL), 25 µL of master mix (dNTP, MgCl2, 10X buffers, Taq DNA polymerase), 5 µL of purified DNA (20 ng), and 17 µL of RNase-free water. The amplification reactions involved the following stages: one denaturation cycle at 94°C for 10 minutes and 40 cycles of amplification at 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute, followed by one elongation cycle at 72°C for 10 minutes. Next, 5 µL of the amplified result was run on a 1% agarose gel in 0.5x TBE, stained by ethidium bromide, and visualized by UV light photography. Next, 6 µL of the PCR product was heated for 5 minutes at 95°C and cooled on ice for 5 minutes, then loaded onto the acryl amide gel at 2 W for 14 - 16 hours in a cold room, and the gel was then silver-stained. SSCP assays were repeated at least two times for each isolate (16).
3.6. Sequencing
PCR products were purified, and samples of the quinolone-resistance-determining region (QRDR) of the gyrA gene amplicons, with different PCR-SSCP patterns from ciprofloxacin resistance and sensitive isolates, were sequenced with the applied biosystems 377 automated sequence procedure (ABI PRISM™ Dye Terminator). All post-run analyses were performed using MEGA4, MEGA5, and ClustalW. ClustalW is a multiple-sequence alignment program and MEGA is software for conducting automatic and manual sequence alignments. Each sequence was compared both with the control strain sequences and with the published gyrA gene sequence in GenBank.
4. Results
Forty-one isolates were identified by phenotypic and molecular tests. The frequency of isolates were as follows: M. fortuitum, 27 cases; M. gordonae, 10 cases; M. abscessus, two cases; and M. conceptionense and M. smegmatis, one case each (Table 1). The susceptibility of isolates to ciprofloxacin concentrations was determined by agar dilution. All isolates except for M. abscessus were sensitive to concentrations of 1 µL/mL, 2 µL/mL and 4 µL/mL of ciprofloxacin (MIC > 1 µL/mL) (Table 2). All clinical isolates and all environmental isolates except for M. abscessus were sensitive to ciprofloxacin. Based on the PCR-SSCP results for the QRDR of the gyrA gene, two ciprofloxacin-resistant M. abscessus isolates had four similar bands, which were different from Mycobacterium fortuitum and M. gordonae isolates. Mycobacterium fortuitum also showed four bands, but the band size and positions were different from M. abscessus, and M. gordonae had three bands (Figures 1 and 2).
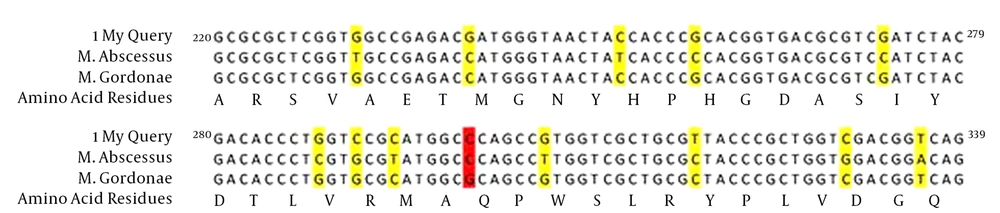
Each Mycobacterium species had an equal and specific PCR-SSCP pattern. The PCR-SSCP patterns of clinical and environmental M. fortuitum isolates with four bands were similar, and the clinical and environmental M. gordonae isolates with three bands were also similar. The amino acid sequence of M. abscessus resistant to ciprofloxacin was 100% similar to M. abscessus ATCC 19977 (HQ324097.1), but the nucleotide sequence was different at 12 points: 231 (T→G), 240 (C→G), 252 (T→C), 258 (C→G), 273 (C→G), 288 (C→G), 291 (G→C), 294 (T→C), 306 (T→G), 318 (C→T), 330 (G→C) and 336 (A→T). Since the amino acid sequences of M. abscessus and M. gordonae ATCC 14470(AJ564389.1) were completely similar, their nucleotide sequences were also compared and were 96% similar (Figure 3).
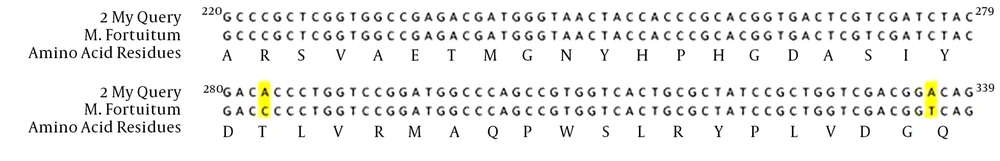
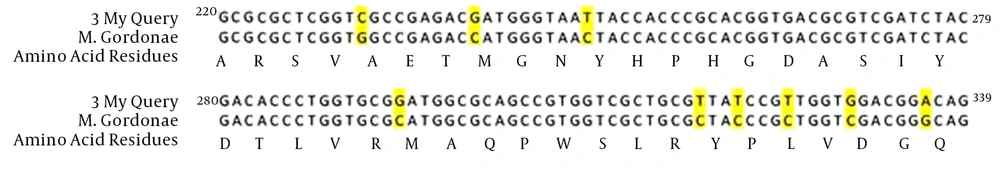
The amino acid sequence of M. fortuitum sensitive to ciprofloxacin was 100% similar to that of M. fortuitum ATCC 6841(AJ564392.1), but the nucleotide sequence was 98% similar and had two alterations, at positions 283 (A→T) and 336 (T→A) (Figure 4). The amino acid sequence of M. gordonae sensitive to ciprofloxacin was 100% similar to M. gordonea ATCC14470 (AJ564389.1), but the nucleotide sequence was 92% similar and had nine alterations: 231 (G→C), 240 (C→G), 249 (C→T), 294 (C→G), 318 (C→T), 321 (C→T), 325 (C→T), 330 (C→G), and 336 (G→A) (Figure 5). The gyrA gene sequences studied were deposited on the national center for biotechnology information website as follows: M. fortuitum (gene name: EsfahanMyco-Fort-GyrA, accession number: LC074879.1), M. abscessus (gene name: EsfahanMyco-Abs-gyrA, accession number: LC074880.1), and M. gordonae (gene name: EsfahanMyco-Gord-gyrA accession number: LC074881.1).
| Mycobacterium Species | Clinical Isolates | Environmental Isolates | Total |
|---|---|---|---|
| M. fortuitum | 15 | 12 | 27 |
| M. gordonae | 5 | 5 | 10 |
| M. abscessus | NA | 2 | 2 |
| M. smegmatis | NA | 1 | 1 |
| M. conceptionense | NA | 1 | 1 |
| M. fortuitum ATCC 49403 | Positive control | NA | Positive control |
Abbreviations: ATCC, American type culture collection; NA, not available.
| Source of Isolate and Mycobacterium Species | Number | Minimum Inhibitory Concentration, µg/mL | Percentage of Inhibition | ||
|---|---|---|---|---|---|
| 4 µg/mL | 2 µg/mL | 1 µg/mL | |||
| Clinical | |||||
| M. fortuitum | 15 | < 1 | 100 | 100 | 100 |
| M. gordonae | 5 | < 1 | 100 | 100 | 100 |
| M. fortuitum ATCC 49403 | Positive control | < 1 | 100 | 100 | 100 |
| Environmental | |||||
| M. fortuitum | 12 | < 1 | 100 | 100 | 100 |
| M. gordonae | 5 | < 1 | 100 | 100 | 100 |
| M. abscessus | 2 | > 4 | 24.80 | 30.80 | 32.80 |
| M. smegmatis | 1 | < 1 | 100 | 100 | 100 |
| M. conceptionense | 1 | < 1 | 100 | 100 | 100 |
5. Discussion
Over the past several years, nontuberculous mycobacteria (NTM) have been reported as some of the most important agents of infection in immunocompromised patients. M. abscessus, M. fortuitum, and M. chelonae are the most important opportunistic pathogens in humans (17, 18). Strategies for the treatment of TB are different from those for NTM, in that the latter is often resistant to anti-tuberculosis agents (2). On the other hand, NTM show variable susceptibility patterns in different geographical regions of the world. Therefore, identification of NTM and determination of drug sensitivity is crucial. Conventional phenotypic methods, such as culture, are time-consuming because of the slow-growing nature of mycobacteria (19). Molecular methods, such as multiplex PCR, targeting many different genes simultaneously, have been used to detect and identify MTC and NTM in routine diagnostic laboratories (20). Mokaddas developed a multiplex PCR targeting the oxyR-ahpC and rpoB genes for the direct identification and differentiation of clinical MTC or NTM isolates in primary cultures (19).
Of the 41 isolates identified by phenotypic and molecular tests, the frequencies were as follows: M. fortuitum, 27 cases; M. gordonae, 10 cases; M. abscessus, two cases; M. conceptionense, one case; and M. smegmatis, one case. All isolates except for M. abscessus were sensitive to all three concentrations (1 µL, 2 µL and 4 µL) of ciprofloxacin (MIC >1 µL/mL). Vacher et al. (21) analyzed 130 isolates of Mycobacterium for susceptibility to grepafloxacin, ofloxacin, and ciprofloxacin. The MIC of the fluoroquinolones was defined using the agar dilution method. Different mycobacterial species showed different degrees of susceptibility to three antibiotics, and the activity of antibiotics against rapidly growing mycobacteria, such as M. fortuitum, was very good. This is consistent with our results. In 2010, Gayathri (22) evaluated rapidly growing mycobacteria drug sensitivity to antibiotics, including ciprofloxacin. Of 148 isolates, 76% were susceptible to ciprofloxacin. A 1996 study by Sanchez-Carrillo et al. (23) on 64 NTM (40 rapidly growing and 24 slowly growing) compared the activity of several antibiotics, such as ciprofloxacin, that had high activity against rapidly growing NTM and good activity against slowly growing ones.
In that study, M. gordonae (slow-growing) were susceptible to ciprofloxacin. In 2009, Rafi et al. (24) evaluated the susceptibility of M. tuberculosis and NTM to two mycobacterial agents (ciprofloxacin and ofloxacin), and the findings demonstrated that ciprofloxacin could be effectively used against TB and NTM. This is similar to the results of the present study. Due to the sensitivity of environmental NTM isolates (except for M. abscessus) and clinical isolates, including M. fortuitum and M. gordonae, to ciprofloxacin, this antibiotic should be regarded as a primary drug in the treatment of these infections. However, it is necessary to determine the < 1 MIC of the concentrations of ciprofloxacin. The sequences of a conserved site in the A subunit of DNA gyrase corresponding to the QRDR were established for NTM species and then compared. The nucleotide sequences were highly conserved, probably because of the essential function of the gyrase, and they clearly differentiated one species from another (13). Furthermore, a domain of the N-terminal part of the A subunit is highly conserved among prokaryotes (25). This domain contains the catalytic site of DNA gyrase. In other words, from residues 67 - 106 in the numbering system used in E. coli, this domain contains the QRDR, which is supposed to be the site of interaction between the A subunit of gyrase and quinolones (26).
Quinolone-resistance-determining region could be involved in intrinsic quinolone resistance in mycobacteria. We compared nucleotide and amino acid sequences of QRDRs from NTM species with different levels of susceptibility to quinolones. The detection of missense mutations at positions 83, 84, and 87 in gyrA is a quick and efficient test for molecular identification of fluoroquinolone resistance in mycobacteria (27, 28). Guillemin et al. (14) evaluated the correlation between quinolone-resistant patterns and sequences in DNA gyrase in 14 mycobacterial species. With regard to MICs, the species could be organized into three groups: resistant, moderately susceptible, and susceptible .Peptide sequences of the QRDR of gyrA were identical in all of the species, except for the amino acid at position 83. This suggests that this amino acid is implicated in the observed differences of quinolone susceptibility within the mycobacteria.
In this study, the peptide sequences of resistant M. abscessus species were similar to those of M. abscessus ATCC 19977, which is resistant to ciprofloxacin. Pitaksajjakul et al. (29) studied gyrA/B mutations in 92 fluoroquinolone-resistant TB isolates in Thailand. There were 70 isolates with point mutations associated with ofloxacin resistance. The other 22 isolates had no mutations in either the gyrA or the gyrB QRDR. In another study, Pitaksajjakul et al. (30) evaluated 35 fluoroquinolone-resistant MTB isolates that were amplified using PCR. DNA sequencing and SSCP were further utilized for characterization of the mutations in the QRDR of the gyrA/gyrB genes. On the DNA sequencing, 60% exhibited single-point mutations at different positions, and there was one novel mutation in the gyrA gene and Asp495Asn in the gyrB gene. Forty percent of the isolates had no mutation.
In our study, there was no difference in the nucleotide sequences of resistant strains to the standard nucleotide sequence. SSCP was performed on 35 fluoroquinolone-resistant MTB isolates for mutations of gyrA/gyrB amplicons, and five different SSCP patterns were obtained. For each fluoroquinolone-resistant and fluoroquinolone-sensitive MTB isolate, the SSCP patterns, in contrast to our results, were indistinguishable on repeated analyses (30). In some Mycobacterium types, point mutations in the drug-binding locations occur, maintaining their protein function, which leads to the bacteria becoming resistant to the drug, and therefore the protein structure is preserved. A conservation type of mutation happens in these bacteria. The point mutation in the nucleotide sequence means that the changes happen on the nucleotide surface, but the codons translate to an amino acid, so we can say that a polymorphism has occurred. In this study, PCR-SSCP was used to evaluate mutants resistant to ciprofloxacin. The PCR-SSCP patterns of mutated ciprofloxacin-resistant isolates were clearly differentiated from the PCR-SSCP patterns of those sensitive to ciprofloxacin.