1. Background
Helicobacter pylori, the most common pathogenic bacterium in the human stomach, is a microaerophilic, Gram-negative organism involved in peptic ulcer disease and gastric cancer (1). Many virulence factors are involved in the pathogenicity of H. pylori, including urease, vacuolating cytotoxin (a product of the vacA gene), and the immunogenic protein CagA, encoded by the cytotoxin-associated gene A (cagA). While the vacA gene is present in all strains of H. pylori, cagA is not (2). The cagA gene is a virulence gene located in the cag pathogenicity island of the bacterial genome, and is frequently associated with more severe clinical outcomes (3-5). The cagA gene encodes proteins that increase the virulence potencies of strains, such as by increasing host-cell cytokine production and altering protein tyrosine phosphorylation (6, 7).
Many investigators have demonstrated that H. pylori possesses a remarkable degree of genetic diversity, closely related to its epidemiological and pathological characteristics and dynamics of transmission. Various methods have been employed for H. pylori genotyping, such as pulsed-field gel electrophoresis (PFGE), PCR-based randomly amplified polymorphic DNA (RAPD) fingerprinting, and hybridization with specific probes (8-10). By using restriction enzyme EcoRI and the PFGE method, a 97% improvement in H. pylori genotyping has been described (11). However, the PFGE method is time-consuming and requires a unique apparatus to handle a large number of clinical specimens. RAPD-PCR, on the other hand, has been reported to be a valuable method for the study of H. pylori genetic diversity and transmission (12). In comparison to PFGE, RAPD-PCR exhibits reasonable speed, cost, and efficiency. However, reproducibility is the main problem with this method. Recently, a novel peptide nucleic acid probe for the specific detection of H. pylori in gastric biopsy specimens has been introduced (13). In this study, specific probes were used for the differentiation of H. pylori from other microorganisms. However, the sensitivity and specificity of this method were not compared with nucleic acid sequencing as a gold standard method.
PCR-RFLP analysis is another molecular method that has been used to investigate H. pylori genes, especially those that encode proteins in relation to bacterial pathogenicity, such as cagA and vacA, and urease structural and accessory proteins (14-18). However, the accuracy of the data provided by the PCR-RFLP method should be verified by a gold standard method when differentiating clinical isolates of H. pylori.
2. Objectives
In this study, we employed PCR-RFLP to investigate the genetic diversity of cagA as a virulence gene among H. pylori isolates recovered from antral biopsies of Iranian patients with stomach symptoms. The results were compared with nucleotide sequencing analysis as a gold standard method, to identify any changes in nucleotide sequences in cagA. The associations between the clinical status of the patients and the different genotypes were also investigated.
3. Patients and Methods
3.1. Patients
In this cross-sectional study, a total of 161 patients presenting to the endoscopy section at the Motahhary clinic, affiliated with Shiraz University of Medical Sciences (SUMS) in Shiraz, Iran, from July to December 2012, were enrolled in this study. Based on previous studies in the same area, the prevalence of H. pylori infection in patients with gastritis is 50% - 60%. Therefore, the sample size was derived with a 95% confidence interval and an estimate error of 7% - 8%. The calculated total sample size for the study was 160 gastric biopsy specimens. The patients had different gastric symptoms, such as stomach pain, nausea, dyspepsia, and gastritis. There were 61 males (37.9%) and 100 females (62.1%) in the study, with an age range of 16 - 80 years. The exclusion criteria were the use of antibiotics or proton pomp inhibitors within two weeks prior to endoscopy, and previous gastric surgery. This study was approved by the ethical committee at SUMS, and all participants provided written informed consent.
3.2. Isolation and Identification of Helicobacter pylori Strains
Two antral biopsy samples were taken from each patient during endoscopic procedures for rapid urease test, gram staining and H. pylori culture for isolation and identification. The presence of urease produced by H. pylori was determined using modified rapid urease agar medium (19). Briefly, one biopsy sample from each patient was inoculated into urea agar medium immediately after collection, and incubated at room temperature. A positive or negative reaction was determined on the basis of a change in color from yellow to red after a maximum period of two hours. Biopsy samples were also cultured on modified Columbia agar plates (Merck, Germany) containing 10% lysed horse blood, and incubated under microaerobic conditions in an anaerobic jar at 37°C for 2 - 3 days, as previously described (20). The organisms were identified as H. pylori by colony morphology, gram staining, and positive oxidase, catalase, and urease tests. Positive cultures were harvested and stored in sterile distilled water at -70°C for further investigations.
3.3. DNA Extraction
The genomic DNA of the H. pylori isolates was obtained using the boiling extraction method. Briefly, 300 µL of thawed isolated H. pylori samples were incubated at 95°C for 10 minutes. The samples were then centrifuged at 14,000 g for 10 minutes and the supernatants were stored at -20°C until being used as PCR templates. DNA concentration was assessed by spectrophotometry (NanoDrop ND-1000, USA).
3.4. Detection of H. pylori and cagA Gene by PCR
To confirm H. pylori species, a set of published primers was used to amplify the 294 bp fragment of the glmM gene, which encodes phosphoglucosamine mutase (21). PCR was performed in a total volume of 25 µL containing 10 ng of genomic DNA from the H. pylori culture, 200 µM of dNTP, 1.5 mM of MgCl2, 0.5 µM of each primer, and 1.25 U of Taq polymerase. The PCR condition for amplification was performed with a one-cycle initial denaturation step at 93°C for 5 minutes, followed by 35 cycles at 93°C for one minute, 55°C for one minute, and 72°C for one minute. A final extension was performed at 72°C for five minutes.
To detect the presence of the cagA gene, primer cagA F [5’-AGT AAG GAG AAA CAA TGA-3’] and primer cagA R [5’-AAT AAG CCT TAG AGT CTT TTT GGA AAT C-3’] were used to amplify a 1320-bp band of the target gene (18). The PCR mixture consisted of 200 µM of dNTPs, 1.5 mM of MgCl2, 0.5 µM of each primer, 1.25 U of Taq DNA polymerase, and 20 ng of DNA template in a 50 µL final volume. Negative and positive controls were also included in the PCR assay. Thermal cycling was set at a one-cycle initial denaturation at 94°C for 2 minutes, 35 cycles of 45 seconds for denaturation at 94°C, 45 seconds for annealing at 50°C, and 45 seconds for extension at 72°C, with a final extension at 72°C for 5 minutes.
The PCR products were electrophoresed on agarose gel, and amplified DNA was stained with ethidium bromide and observed under gel documentation (UVitec, England). A 100-bp DNA ladder was used as a molecular size marker.
3.5. RFLP Assay
The cagA gene amplicons were digested by HinfI as an endonuclease enzyme, according to the manufacturer’s instructions. The site recognized by this enzyme is 5’ G*A N T C 3’. For this purpose, 10 μL of each PCR product of the cagA gene was mixed with 1 U of HinfI restriction enzyme (Fermentase, Fermentas, Lithuania) and 1 μL of 10X restriction buffer, then incubated at 37°C for 3 hours. The digested products were then electrophoresed on 2% agarose gel, stained with ethidium bromide, and observed under gel documentation (UVitec, England). A 100-bp DNA ladder was used as a marker for molecular size.
3.6. Sequencing of cagA Products
After RFLP analysis, one strain of each genotype of cagA PCR product was randomly selected and sequenced. For sequencing, the cagA PCR products were first purified with a column-based purification kit (Millipore), then sequenced by the Sanger method using an ABI 3730XL instrument (Sequetech Corp. CA, USA). The recognition sites of the HinfI enzyme within the sequence region were analyzed with the Serial Cloner V2.5 program.
3.7. Sequence Analysis of cagA PCR Products
The sequenced cagA genotypes were aligned with GenBank accession number AE000511 (corresponding to nucleotides 579907 - 581241 of H. pylori 26695) as a reference. The homology of the DNA sequences to the published sequences was determined by using the BLAST window on the National center for biotechnology information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/).
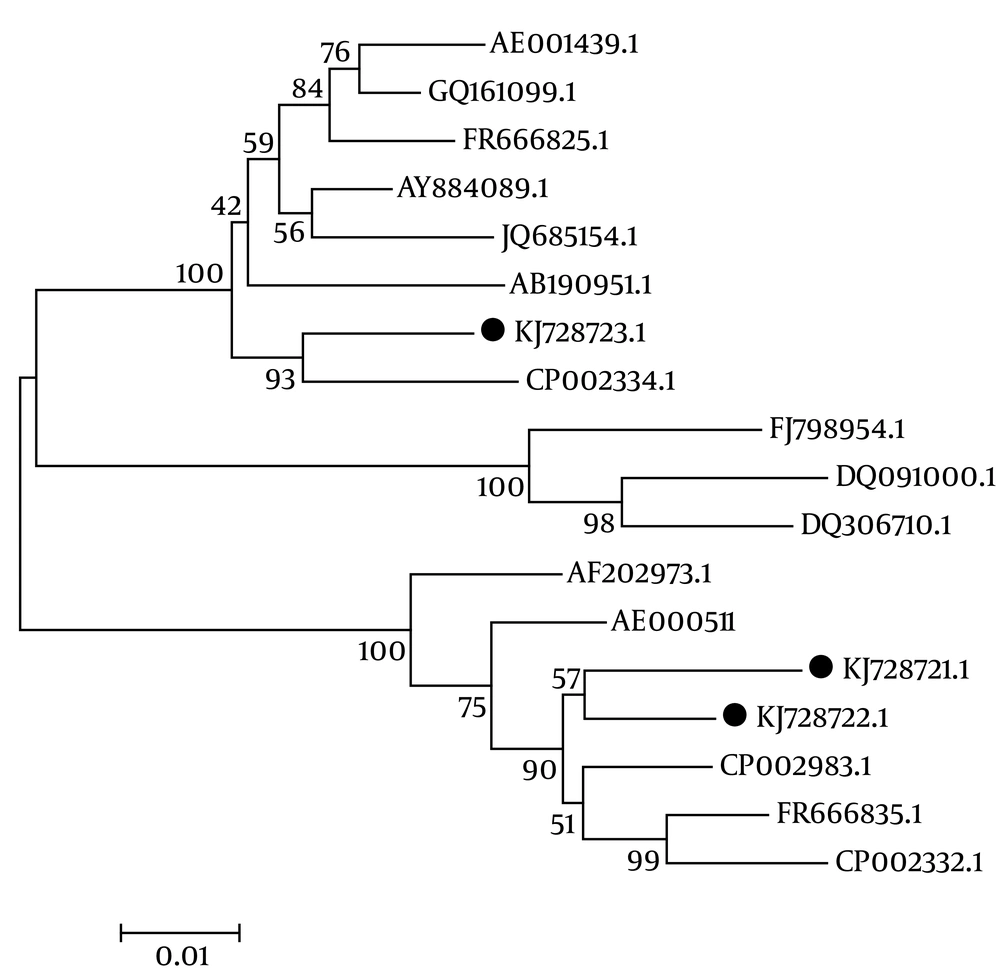
Three sequenced genotypes (KJ728721.1, genotype I; KJ728722.1, genotype II; and KJ728723.1, genotype III) were aligned with 15 sequences available in the GenBank database, using the CLUSTAL W method (accession numbers: AE000511.1, 26695; AE001439.1, J99; JQ685154.1, J166 USA; CP002983.1, SNT49 USA; FR666835.1, USA; FR666825.1, UK; CP002332.1, Gambia; CP002334.1, Lithuania; GQ161099.1, China; DQ306710.1, China; AY884089.1, Korea; AB190951.1, Japan; DQ091000.1, Japan; AF202973.1, Japan; and FJ79854.1, Vietnam).
The phylogenetic analysis was performed using Mega 4 software (Beta MacOSX version) following the recommended instructions. The phylogenetic tree was constructed via the neighbor-joining method following 500 bootstrap surveys.
3.8. Statistical Analysis
Statistical analysis of the data was performed with Fisher’s exact test using SPSS version 17.0 (Chicago, IL, USA) for Windows, and a P-value of less than 0.05 was considered significant.
4. Results
4.1. Isolation of Helicobacter pylori From Biopsy Specimens
Overall, 61 out of 161 antral biopsy specimens were positive for H. pylori according to the rapid urease test and culture. All of the isolates were confirmed by the glmM gene PCR assay, as well. With regard to the endoscopic examinations, there were diagnoses of gastritis (n = 47, 77%), duodenitis (n = 7, 11.5%), and duodenal ulcers (n = 7, 11.5%).
4.2. PCR Amplification and Restriction Fragment Length Polymorphism Analysis of the cagA Gene
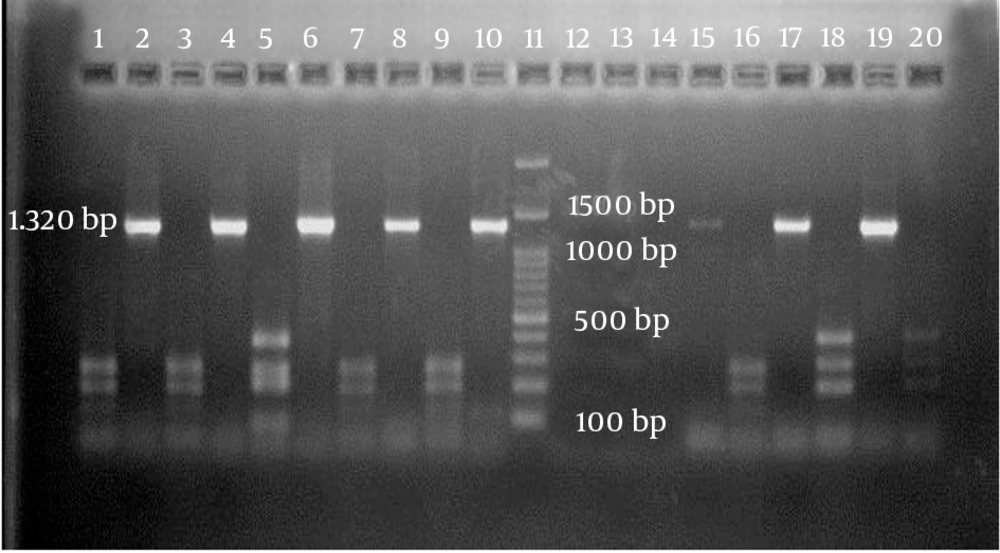
Using a specific primer, a 1320-bp fragment of the cagA gene was detected in 24 out of 61 isolates (39.3%). Of those, 17 isolates belonged to patients with gastritis (70.9%), two were in patients with duodenitis (8.3%), and five were in patients with duodenal ulcers (20.8%). The distributions of H. pylori cagA-negative strains isolated from patients with gastritis, duodenitis, and duodenal ulcers were, respectively, n = 30, 81.1%; n = 5, 13.5%; and n = 2, 5.4%. When the RFLP analysis was performed on the 24 cagA-positive isolates using HinfI restriction endonuclease, three different genotypes of cagA were determined on agarose gel electrophoresis (Figure 1).
Genotypes I and II had three distinct DNA fragments with different sizes. Genotype III, on the other hand, revealed four DNA fragments on agarose gel electrophoresis. Among all of the H. pylori cagA-positive isolates, the prevalence of genotype I was significantly higher than the other two genotypes, at 62.5% versus 25% and 12%, respectively (P = 0.014). The associations of each of the cagA genotypes with the gastroduodenal diseases are shown in Table 1.
| cagA Genotypes | No. of Isolates From Gastroduodenal Disease | Total | ||
|---|---|---|---|---|
| Gastritis | Duodenitis | Duodenal Ulcer | ||
| I | 12 (70.6)b | 0 (0) | 3 (60) | 15 (62.5) |
| II | 5 (29.4) | 0 (0) | 1 (20) | 6 (25) |
| III | 0 (0) | 2 (100) | 1 (20) | 3 (12.5) |
| Total | 17 | 2 | 5 | 24 |
aValues are expressed as No (%).
bP = 0.014.
4.3. Sequence Analysis of cagA PCR Products and BLAST Query
Table 2 indicates the identities of cagA gene nucleotides, determined by PCR direct sequencing, and the deduced amino acid sequences among three H. pylori isolates and H. pylori 26695, using the NCBI protein BLAST query.
| Comparison of Strains | Identity of Nucleotide Sequence | Identity of Amino Acid Sequence |
|---|---|---|
| I with H. pylori 26695 | 1293/1335 (97) | 420/440 (95) |
| II with H. pylori 26695 | 1302/1335 (98) | 428/440 (97) |
| III with H. pylori 26695 | 1231/1349 (91) | 388/444 (87) |
| I with II | 1297/1335 (97) | 424/440 (96) |
| I with III | 1211/1349 (90) | 378/444 (85) |
| II with III | 1226/1349 (91) | 387/444 (87) |
aThe values are expressed as No (%).
4.4. Submission of Three Genotypes to the NCBI Database
Three sequenced cagA genotypes were submitted to the NCBI database. Of the three cagA-positive H. pylori strains, two were from non-ulcer disease (NUD) patients with gastritis (GenBank accession numbers KJ728721 and KJ728722), and one was from a duodenal ulcer (DU) patient (GenBank accession number KJ728723).
4.5. PCR Product Sequence Analysis of Three cagA Genotypes
Each of the sequenced cagA genotypes were entered into the serial cloner V2.5 program, which revealed the actual fragment size digested by the HinfI restriction enzyme (Table 3).
| Strain Group | Fragment Length | Sum |
|---|---|---|
| Reference strain H. pylori 26695 | 274, 249, 208, 199, 127, 70, 58, 43, 43, 36, 15, and 13 | 1335 |
| Genotype I | 273, 249, 208, 199, 127, 70, 58, 43, 43, 36, 15, and 13 | 1335 |
| Genotype II | 376, 274, 208, 199, 70, 58, 43, 43, 36, 15, and 13 | 1335 |
| Genotype III | 375, 266, 223, 207, 102, 79, 43, 36, and 15 | 1347 |
aThe values’ unit is bp.
4.6. Analysis of PCR Product Sequences of Three cagA Genotypes in Comparison With the Helicobacter pylori Reference Gene
The nucleotide sequences of three cagA genotypes were compared to nucleotides 579907 - 581241 of H. pylori 26695, which indicated that there were some base-substitution events within the cagA gene of the strains isolated from the patients. Some of these changed the sequence bases to the non-cut sites of HinfI; therefore, three genotypes (I, II, and III) of cagA were detected among the isolates. In genotype II, there was a base substitution of G1193A that provided a region not cut by HinfI. Interestingly, in genotype III, the base substitutions of G208A, AA337-8GT, C539T, G548A, T551C, and G1219A induced non-cutting sites by HinfI. On the other hand, the base substitutions of GA369-70AC and G801T induced new cutting sites by the enzyme. No insertions or deletions were found within the cagA region in genotypes I and II. However, two base deletions and 14 base insertions were detected in genotype III. The deduced amino acid sequences could be determined without a stop codon, and there were also some differences in the nucleotides and the amino acid sequences among the three cagA genotypes.
Alignment of the nucleotide sequences of the three isolated genotypes, with H. pylori 26695 as a reference strain, revealed that in genotype III (KJ728723.1), 12 nucleotides had been inserted (GAG AGC CTA CTG) in positions nt 626 – 637. When the sequence of genotype III was aligned with 15 additional H. pylori strains available in the GenBank database, the same sequences were detected among six strains with the following accession numbers: GQ161099.1, China; AE001439.1, J99; FR666825.1, UK; JQ685154.1, J166 USA; AY884089.1, Korea; and AB190951.1, Japan.
The phylogenetic analysis revealed a sequence disparity among the three isolated genotypes, as depicted in Figure 2. This result demonstrated that two isolates in this study (genotype I = KJ728721.1, genotype II = KJ72871.1) had the same diversity, as they were located close together in the same branch. Another isolate (genotype III = KJ728723.1) showed a different phylogenetic pattern, as it moved to another taxa on a completely separate branch.
5. Discussion
In the present study, we employed PCR-RFLP to investigate the genotypes of the cagA gene in H. pylori strains isolated from antral biopsies of patients with stomach symptoms. The cagA gene was detected in 39.3% of H. pylori-positive isolates. However, no significant association was found between the presence of the cagA gene and upper gastrointestinal disorders in the patients infected with H. pylori. Based on different geographical regions, the prevalence of cagA-positive strains among the Iranian population is between 35% and 94% (22-25). In a previous study in Shiraz, 47.7% of H. pylori isolates were cagA-positive, which was higher in patients with UD than in those with NUD. However, no significant difference was detected in the prevalence of the cagA gene between the two groups (22). The variability of cagA status might be due to the different PCR primer sets used in different studies. There may be several distinct forms of the cagA gene, with an uneven geographical distribution. Such differences in cagA genotypes can serve as markers for differences in virulence among cagA-positive strains.
Using the HinfI restriction enzyme, PCR-RFLP revealed three distinct genotypes of cagA among the H. pylori strains according to base size and number of bands (fragments) in agarose gel, which confirmed that there is genetic diversity in the cagA gene of the isolates. Genotypes I and II of cagA were predominant in patients who had gastritis, while genotype III was found in three patients with duodenitis and duodenal ulcers. In a previous study using the same restriction enzyme, two genotypes, α (no cut) and β (3 bands), with no significant association with a specific clinical outcome, were identified (18). Since the PCR products were not sequenced in that study, it is not obvious whether genotype β is identical to genotypes I or II in our study.
Takamura et al. (26) employed PCR-RFLP for genotyping the H. pyloricagA gene isolated from paraffin-embedded sections of gastric cancer. The study populations were Brazilian and Japanese patients. Analysis of PCR-RFLP for the cagA gene showed that the prevalence of the east Asia subtype was significantly higher in the Japanese subjects than in the Brazilians (26), and the authors concluded that this genotype contributes to the progression of gastritis in the region.
In general, the PCR-RFLP method is a useful tool for genotyping the H. pylori cagA gene among clinical isolates. However, this method is limited to the detection of mutations only at the restriction sites of the enzymes, even if many other regions differ throughout the complete genome. The PCR direct sequencing method has been applied in the typing of H. pylori clinical isolates (27, 28). This method can provide a tool for analyzing the nucleotide alignment within a gene; therefore, it has some advantages over other methods that analyze only restriction-site changes in a single gene, such as PFGE and PCR-RFLP.
The results of the RFLP analysis resolved by agarose gel electrophoresis were nearly identical to the predicted fragments, based on the nucleotide sequence data. However, the DNA fragments below 70 bp, as predicted from the nucleotide sequence, were too small to be seen on agarose gel. Since the agarose gel we used in this study was not able to discriminate between DNA fragments with a few nucleotides of different sizes, fragment overlap was likely.
Analysis of the nucleotide sequences indicated that there were some base-substitution events within the cagA gene of the strains isolated from the patients. Some changed the sequence bases to the non-cut sites of HinfI, and vice versa. Therefore, three genotypes (I, II, and III) of cagA were detected among the isolates. Some base substitutions, insertions, or deletions induced new amino acid sequences throughout the cagA gene; for instance, substitution of the codon GAA with GGT in nucleotide positions 337 and 338 changed negatively-charged glutamic acid into neutral glycine. Since the majority of the nucleotide alterations occurred in genotype III, it is possible that the severity of disease caused by this genotype is attributed to mutations in the nucleotides. On the other hand, variation in some nucleotides affected the HinfI activity and induced non-cutting sites on the sequence.
Alignment of the three sequenced genotypes of H. pylori cagA PCR products in our study with different H. pylori cagA genotypes, which were submitted to the NCBI GenBank database, revealed 12 inserted nucleotides (GAG AGC CTA CTG) in positions nt 626 – 637 in genotype III. Six of the aforementioned sequences were translated to glycine, glutamic acid, proline, and threonine (GEPT) amino acids in positions aa 209 – 212. GEPT amino acids are located in the amino-terminal domain (D1; aa 24 – 221) of cagA, which activates inflammatory responses via NFkB and prevents apoptosis via the tumor suppressor p53, and this may impact the severity of disease (29).
The phylogenetic analysis revealed that compared to genotype III, the isolated genotypes I and II had much greater similarity to H. pylori SNT49 USA, H. pylori Gambia, and H. pylori 26695. On the other hand, genotype III was much more similar to H. pylori J99 Lithuania, H. pylori Korea, and H. pylori UK. These data indicate that there is diversity among cagA H. pylori strains isolated from different geographical regions, which may impact the severity of disease.
In conclusion, three distinctive H. pylori cagA genotypes were detected in antral biopsies, using the PCR-RFLP method. Genotype I, which was predominant among the isolates, was significantly associated with gastritis. However, the inserted nucleotides in cagA genotype III, which were revealed by the nucleotide sequencing analysis, may play a role in duodenitis and duodenal ulcers in patients infected with H. pylori. To support the data, more clinical samples from patients with different gastrointestinal symptoms and from different geographic regions need to be tested. Sequencing and comparisons of complete cagA genes isolated from different geographic regions is also recommended.