1. Background
HIV-1 is an etiologic agent and a member of the retroviridae family and lentivirus genus (1). A diagnostic characteristic of this virus is its broad genetic diversity. It also shows rapid turnover and exerts selective immune pressure of the host (2-4). HIV-1 is classified into M, N, and O groups. In 2009, group P was added. The P group, which was isolated from a female Cameroonian patient, is closely related to immunodeficiency viruses found in gorillas (5).
Group M is responsible for most HIV-1 infections globally and is divided into 10 subtypes (A - K). The most common genetic forms of HIV-1 are subtypes B, A, and C. Subtype C is responsible for almost 50% of HIV-1 infections. Within group M, the mean genetic diversity of the Gag gene is 15% and that of the envelope (Env) gene is 25%. There are over 20 types of circulating recombinant forms (CRFs) (6), the most common of which are CRF01-AE and CRF02-AG (7). The aforementioned makes it difficult to detect the virus using nucleic acid techniques.
The increasing spread of HIV-1 highlights the urgent need for improved diagnostic methods to detect the infection at an early stage and stem the spread of this disease. Currently, the major methods used to detect HIV-1 are enzyme linked immunosorbent assay (ELISAs), followed by the Western blot method (8, 9). During the window period, diagnostic tests based on specific antibodies cannot detect the presence of the virus, and serological tests cannot diagnose HIV-1 in infants born to infected mothers because of the active transfer of maternal antibodies to the infant (10). The development and application of rapid and accurate diagnostic tests and advanced molecular techniques are of the utmost importance. Such tests could slow the spread of HIV-1 by speedy detection of infected individuals and products. Once detected, the viral load of infected individuals can be measured to determine their prognosis and draw up a treatment plan, in accordance with international standards.
2. Objectives
The present study describes a sensitive, efficient, and easy method to detect HIV-1. The study utilized new, highly specific primers for the protease gene site, in combination with nested RT-PCR, to detect HIV-1 subtypes that show extreme nucleotide variation. As the Pol gene is a protected region of the virus genome, primers targeting this area were used for the detection of HIV-1.
3. Patients and Methods
The primers were designed, as follows: First, the alignment of approximately 1,000 pol sequences was carried out using the Gen Bank database, and MEGA4 software (www.megasoftware.net) was used to identify protected regions. The reference sequences used were Ref.A1.AU.03.PS1044_Day0.DQ676872, Ref.B.FR.83.HXB2_LAI_IIIB_BRU.K03455, Ref.C.ZA.04.SK164B1.AY772699, and Ref.D.CD.83.ELI.K03454). Gene Runner software (http://www.generunner.com), an Oligo analyzer (http://eu.idtdna.com), and Oligo6 (www.oligo.net) were used to design the primers. The final primers that were selected were analyzed using BLAST software (ftp://ftp.ncbi.nlm.nih.gov/blast/db/). The primer sequences were sent to the Alpha Institute of Canada for verification.
All the primers used in the present study were new and original products developed for use in the present study. The present study was descriptive. The samples were collected from a population of patients infected with HIV-1. In this study 25 serum samples of patients under treatment and also 10 positive and 10 negative control samples were studied. The study was approved by the bio-safety and medical ethics committee of Tarbiat Modares University. All the patients signed informed consent forms. Using a 0.002 error rate and a confidence level of 95%, the number of samples selected was 25. All samples had been previously confirmed to be positive using the Western blot technique, and all were from HIV-1 patients who were receiving treatment.
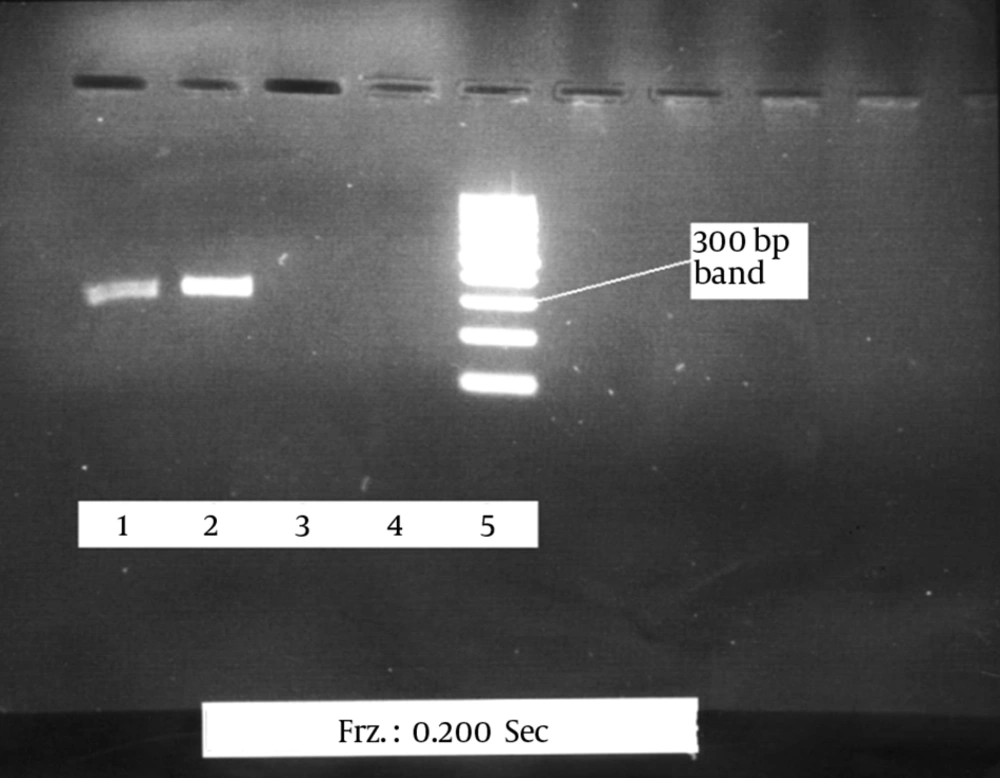
Approximately 10 mL of peripheral blood was collected from each HIV-1 patient. Viral RNA was extracted using the RNA extraction kit (QIAGEN, Hilden, Germany). The extracted RNA was immediately converted into cDNA. The first and second rounds of PCR were done. The product of the first round of PCR was 586 bp, and the product of the second round was 325 bp. A 100 bp marker (DNA Ladder, Fermentas, Canada) was used. To verify the accuracy of the final product, the 25 HIV-1 positive samples were sent the Alpha Institute of Canada to be sequenced. Nested RT-PCR was optimized to detect HIV-1 using three positive samples. The results were then confirmed using the Western blot method. To optimize the PCR reaction to detect low levels of the virus in the samples and to obtain the best possible band, using the most appropriate materials and reaction temperatures to create a strong and pure band.
The data and results of the positive, negative, and control samples were analyzed using Pearson’s correlation, regression, and ANOVA analyses and SPSS17 software.
4. Results
The efficiency of the proposed method was tested on 25 plasma samples from patients infected with HIV-1, 10 positive serum samples, and 10 negative serum samples. The 325 bp band of the protease gene was clearly observed in all the positive samples after electrophoresis. No band was detected in the negative samples, and there were no nonspecific bands or smears (Figure 1).
The proposed method correctly identified HIV-1 in 23 of the 25 samples. The results of the sequencing of the positive cases further confirmed the accuracy of the proposed method (Table 1). Given that the viral load in the HIV-1 patients who were receiving treatment was low, the detection rate (23 of 25 cases) is satisfactory.
| Primer | Sequence |
|---|---|
| External reverse | TGCCCTATYTCTAARTCAGATCC |
| External forward | TTAGYCCTATTGARACTGTACCAG |
| Internal reverse | AATATTGCYGGTGAYCCTTTCCATC |
| Internal forward | GCCTGAAAATCCATYCAAYACTCC |
5. Discussion
Since the discovery of HIV-1 in1984 as the etiologic agent of AIDS, much progress has been made in identifying this virus. Serological tests, including antibody detection, have increased the sensitivity and specificity of the tests. HIV-1 antigen detection tests and cultivation of this virus are useful in some clinical procedures. Most cultures, with the exception of those from patients receiving highly active antiretroviral therapy (HAART) therapy, will be positive within 21 days. The sensitivity of this technique (cultivation of HIV) in patients whose blood serum test results are positive is 97%, and the specificity is 100%. The cultivation of HIV is difficult, time-consuming, and less sensitive than PCR. The ELISA method, which is based on the detection of the p24 antigen, is also not useful for the detection of HIV-1 because it has low sensitivity.
Qualitative and quantitative methods can be used for the examination of nucleic acids of a virus. In qualitative methods, the detection of the nucleic acid of the virus is done by RT-PCR. The PCR method can be used to detect the provirus form in peripheral blood monomorphonuclear cells (PBMC) cells. This method is useful for the diagnosis of infants born to HIV-1-infected mothers and is more sensitive than virus cultivation and the p24 antigen (9).
In the present study, the proposed nested RT-PCR method was used to detect HIV-1. RT-PCR is a sensitive technique, and its sensitivity increased using the nested RT-PCR approach. In this study, part of the RT-PCR product was used as a template for the second-round PCR. To avoid amplification of primer dimers and nonspecific products generated in first-round PCR, a series of internal primers were used. The nested primers were internal, as compared to the primers used in the first round of the PCR and generated shorter products. As the sequencing of the second-round PCR primers was different from that of the first-round PCR primers, the primer dimers and nonspecific products produced in the first round were not duplicated in the second round. This created a more specific product for the second-round PCR.
The sensitivity and specificity of the proposed method was greater than those of existing methods. The proposed method can be used to detect viral RNA in serum samples with viral loads of less than 100 copies per mL (11). HIV-1 transmission through blood and blood products during the window period is of great concern. The application of a sensitive, accurate, and rapid method for the detection of HIV-1 in infants born to infected mothers is essential and requires a method that can identify this stage of infection.
5.1. Conclusions
The results indicate that the proposed nested RT-PCR method had high sensitivity and specificity, as compared with previous methods. The proposed method is appropriate for use in diagnostic centers as a supplementary technique. In particular, it may be useful for the detection of infection in infants born to HIV-1 infected and the diagnosis of infections in the window period.