1. Background
Streptococcus pneumoniae colonizes the nasopharynges of approximately 50% of adults as a part of their normal flora (1). This process is a primary step for the interaction of bacteria with host cells (2). Afterward, these bacteria can migrate to the bloodstream and establish disease in the host, causing respiratory tract infections and leading to invasive diseases, such as septicemia and meningitis (3). Recent studies have shown that the presence of pili enhances the ability of the initial bacteria to adhere to epithelial cells (4). Two different pilus islets (PI-1 and PI-2) encoded by S. pneumoniae play a role in the initial host cell’s contact with the respiratory tract (5). A chromosomal rlrA pathogenicity islet with a size of 14.2 kb encodes the PI-1 (6) and contains seven genes: rlrA, a positive transcriptional regulator of the gene cluster for pilus expression, three pilus subunits (rrgA, rrgB, and rrgC), and three sortase enzymes (srtB, srtC, and srtD) (7).
PI-2 is a 7-kb region that consists of five genes that encode two surface proteins (PitA and PitB), a signal peptidase-like protein (SipA), and two sortases (SrtG1 and SrtG2) (5). The PI-2 is composed of polymers of the major pilus protein PitB that contain accessory pilus proteins that serve as adhesins, while both the signal peptidase-related protein SipA and sortase SrtG1 are necessary for the assembly and polymerization of the pilus (8). As previously mentioned, the presence of pili is not only associated with serotype but is related to clonal property as well (9). The molecular typing of S. pneumoniae can be performed using different techniques, such as ribotyping, pulsed-field gel electrophoresis, and DNA analysis using polymerase chain reaction (PCR) (10). DNA analysis based on the BOX element has some of the best discriminatory potential. BOX-PCR is DNA typing procedure for amplifying human genome-like tandem repeat DNA elements (10). This method is a quick and very reproducible technique for quantitating the genotypic variation among pneumococcal isolates. Typing of microbial pathogens in a quick and effective manner is of great importance for microbiology and epidemiology (11). Furthermore, this method enables the analysis of precise band sizes without requiring software (12).
2. Objectives
In view of the importance of pili in pneumococcal infections and introducing new protein conjugated pneumococcal vaccine-like pili in recent studies, this investigation was undertaken in Tehran, Iran, to study the prevalence of pili in pneumococcal strains isolated from patients and normal flora and also to determine their clonal types using BOX PCR.
3. Materials and Methods
3.1. Sample Collection and Identification of the Isolates
We analyzed 162 isolates (92 collected from patients in 15 medical centers in Tehran hospitals and 70 from normal flora samples) for the presence of PI-1 and PI-2 from 2010–2013. Participants who had not completed any antibiotic course of treatment at least six months prior to the sampling were analyzed for the carriage of nasopharyngeal S. pneumoniae. The sources of the clinical isolates were found to be cerebrospinal fluid (CSF) (n = 10), blood (n = 20), eye infections (n = 24), the trachea (n = 5), lung secretions (n = 16), ear infections (n = 6), pleural fluid (n = 3), urine (n = 1), the sinuses (n = 5), ascites (n = 1), and abscesses (n = 1); these results are shown in Table 1.
| BOX Type | Normal Flora No. | Clinical Sample No. | Sources of Clinical Isolates |
|---|---|---|---|
| 1 | 7 | 17 | CSF (2), ear (1), blood (3), eye (9), ascites (1), trachea (1) |
| 2 | - | 1 | Eye (1) |
| 3 | 2 | 2 | Blood (1), pleura (1) |
| 4 | - | 3 | Lung secretions (3) |
| 5 | - | 1 | Pleura (1) |
| 6 | 1 | 1 | Sinuses (1) |
| 7 | 12 | 5 | Abscess (1), eye (2), blood (1), lung secretions (1) |
| 8 | 4 | 9 | Blood (3), eye (2), lung secretions (1), CSF (2), trachea (1) |
Properties of the PI-1 Positive Isolates
All clinical samples were initially assessed for the presence of S. pneumoniae using blood agar plates that were incubated at 37°C. These samples were also sub-cultured for purity before further testing. First, pneumococcal identification was confirmed by colony morphology, α-hemolysis, optochin susceptibility, and bile solubility assays. All pneumococcal isolates were isolated and confirmed using specific primers for the amplification of the lytA gene (13).
3.2. Antimicrobial Susceptibility
In this study, the minimum inhibitory concentrations (MICs) of penicillin, erythromycin, and tetracycline for the 162 isolates were determined using the E test (Liofilchem, Via Scozia, Roseto d. A., Italy) method on Mueller-Hinton agar. All plates were incubated at 37°C for 24 hours and were evaluated according to the guidelines of the clinical laboratory standards international (CLSI) (14).
3.3. DNA Extraction and PCR Protocol
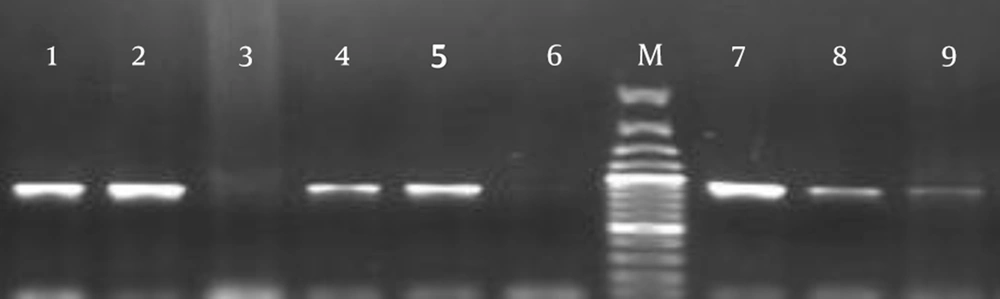
Extraction of bacterial DNA was performed using a DNA purification kit (Roche, Manhiem, Germany) according to the manufacturer’s instructions. Primers were used on the principle of the primary structure of PI-1 and PI-2 for rlrA, rrgC, and sipA, as listed in Table 2 (8, 15, 16). The PCR program was as follows: rlrA, rrgC, and sipA, 30 cycles at 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 1 minute, followed by 5 minutes at 72°C. Each PCR product was purified and directly sequenced for confirmation.
List of Primers Used in This Study
3.4. BOX-PCR
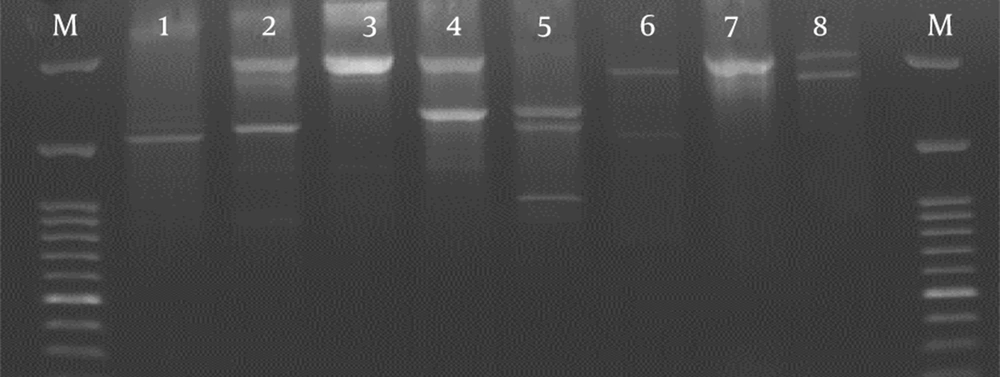
BOX-PCR was carried out using the primer of boxA 5’- CTA CGG CAA GGC GAC GCT GAC G -3’, an interspersed, repetitive DNA sequence disseminated widely among bacterial genomes. The PCR program was designed as follows: predenaturation at 95°C for 7 minutes, 30 cycles each of denaturation at 90°C for 30 seconds, primer annealing at 48°C for 1 minute, chain extension at 65°C for 8 minutes, and a single cycle extension at 65°C for 16 minutes (17).
3.5. Gel Electrophoresis and Pattern Analysis
The amplified products were separated by size on a 1% agarose gel run in 0.5 X Tris-borate-ethylenediaminetetraacetic acid at 100 V for 3 hours. The gel was stained with ethidium bromide (1 mg/mL) and viewed with ultraviolet light. The details were evaluated using a visual examination of the banding patterns.
3.6. Statistical Analysis
All statistical analyses of the links were carried out using the IBM statistical package for the social sciences (SPSS) for the analysis of categorical data. All information was calculated with the fisher’s exact test.
4. Results
4.1. Prevalence of PI-1 and PI-2
Among the S. pneumoniae isolates, 65 (40%) isolates (39 (60%) clinical and 26 (40%) normal flora isolates) were positive for rlrA and rrgC (Figure 2). The most common sources of clinical isolates harboring rlrA and rrgC were found to be eye infections in 36% (n = 14), followed by the blood (n = 8), which were significantly (P < 0.05) higher than any other sites. Other sources included CSF (n = 4), ear infections (n = 1), the pleura (n = 2), the trachea (n = 2), the sinuses (n = 1), abscesses (n = 1), lung secretions (n = 5), and ascites (n = 1) (Table 1). The presence of PI-2 was detected by PCR analysis that used the sipA gene as a marker for this islet; no positive isolate was found in the 162 S. pneumoniae isolates.
4.2. Antibiotic Resistance
The MIC results for penicillin showed that 25% (n = 40) of our isolates were penicillin nonsusceptible (MIC > 2). Seventeen (26%) isolates that contained PI-1 were penicillin nonsusceptible (MIC > 2). Amongst the remaining isolates without PI-1, 23 (23%) were penicillin nonsuceptible. In addition, the MIC for erythromycin and tetracycline revealed that 30 (46%) isolates were resistant to these antibiotics. Eight (12%) isolates with PI-1 were resistant to all three antibiotics, while 8% (8/97) of the isolates without PI-1 were resistant to all three antibiotics (Table 3). Furthermore, the MIC results indicated that 46% of the clinical isolates were resistant to at least one antibiotic.
| Antibiotic Resistance | No. (%) MIC Penicillin > 2 | No. (%) MIC Erythromycin > 16 | No. (%) MIC Tetracycline > 16 |
|---|---|---|---|
| Pilus islet 1 (+) | 17 (26) | 30 (46) | 30 (46) |
| Pilus islet 1 (-) | 23 (23) | 58 (59) | 58 (59) |
Antimicrobial Susceptibility of PI-1 Positive and Negative Isolates
4.3. Clonality of Pilus-Harboring Isolates
The BOX-PCR analysis of 65 isolates carrying PI-1 provided a total of 8 different patterns with two to five bands (Figure 3 and Table 1). Six types (with > 2 isolates) accounted for 98% of the total isolates. Types 1, 7, and 8 contained the highest number with 24, 17, and 13 isolates, respectively. Types 1, 3, 7, and 8 included isolates from the blood and CSF. Type 1 constituted 9/14 (64%) of all strains isolated from eye infections. Some of the types (e.g., 2, 4, and 5) contained only clinical isolates. Other types were found both in clinical and normal flora samples.
5. Discussion
Because they play a major role in adhesion to host cells and are also associated with a number of different virulence mechanisms, the pili of Gram-positive bacteria have recently received considerable attention (4). In particular, the S. pneumoniae pilus was found to be associated with the initial adherence of bacteria to epithelial cells (3) and is regarded as a potential vaccine candidate (18).
As already mentioned in other studies, PI-1 was present in 21% - 27% of clinical isolates depending on the geographic region analyzed (9), but PI-2 is rarely detected in pneumococcal isolates (19). We found that 40% of isolates contained PI-1, which was in agreement with previous findings. A comparison of our results with those of other studies suggested a higher incidence of PI-1 (5, 9). On the other hand, our study demonstrated that none of the isolates contained PI-2. In contrast, Zahner and colleagues identified 21% PI-2 in their isolates (8). Another study also indicated the presence of PI-2 (16%) in global collections (1). This result confirms that the prevalence of PI-2 is related to particular geographic regions and populations; published data have shown PI-2 isolates in global collections are included in the 13-valent pneumococcal conjugate vaccine now being used in an increasing number of countries (8).
PI-1 (rlrA, rrgC) and PI-2 (sipA) genes were used to detect these structures between our isolates. The rlrA gene has been identified as an essential virulence gene in the murine infection model (7). Moreover, other studies have shown that these genes are also essential for expression of the pili (1, 9). Although Basset et al. discussed the limitations of using the detection of rrgC by PCR as a marker for the presence of a pilus islet (6), this may have been too broad a criterion because strains that carry the rrgC gene may lack other genes essential for pilus expression (9). The present study also had the same limitation. The PCR performed to identify the rrgC and rlrA genes indicated that the rlrA islet was present in all isolates. Regarding PI-2, the signal peptidase-related protein SipA is necessary for the assembly and polymerization of the pilus (8). Our isolates lacked this gene, which explains their absence of PI-2.
Our results revealed that the prevalence of PI-1 in clinical isolates was significantly (P < 0.05) higher than normal flora isolates. As suggested in previous studies (4, 20), our results indicated that the pili are important factors in bacterial pathogenesis and disease. Conversely, it seems that strains that are overrepresented in the normal flora would be more likely to acquire the pilus from other strains (6). However, Basset and colleagues demonstrated that the similar frequency of rrgC-positive strains in both normal flora and bacteremic isolates denotes that the pilus does not play an important role in virulence (6). In addition, they suggested that the pilus does not represent an important virulence factor for invasive diseases in humans (6).
However, our results showed that the prevalence of PI-1 was significantly higher in blood samples as well as in pneumococcal isolates from eye infections. We also proved a connection between the presence of a pilus islet and resistance to antimicrobials in clinical collections. Most of the clinical isolates were resistant to at least one antibiotic. However, we did not observe this same association between the pilus islet and normal flora collections of S. pneumoniae. In addition, our results indicated that the prevalence of resistance to the three antibiotics we tested was significantly higher in pilus-harboring isolates than it was in the isolates that lacked it. Other studies (8, 9) have also concluded a similar relationship between the rlrA and antimicrobial resistance.
Our BOX PCR result showed that the isolates harboring a pilus had four main types. This result is inconsistent with a previous study, which showed that the presence of a pilus locus is a clonal property among invasive pneumococcal isolates (6). In all, the data obtained in this study revealed that the rate of PI-1 in our society is high. The fact that the majority of isolates with PI-1 were collected from clinical samples (especially in invasive isolates) suggests the role of this structure in infection. According to the BOX PCR typing, the strains were also arranged in clonal groups.