1. Background
Staphylococcus epidermidis is the most common bacteria isolated from the human skin and is the most persistent bacteria in some body parts. Staphylococcus epidermidis, which is part of the human flora, is also recognized as an opportunistic pathogen and causes nosocomial infections associated with medical devices (1). More information about bacterial physiology not only under infectious conditions but also when S. epidermidis is in a symbiosis condition could help evaluate the therapeutic strategies in the case of infection. It has been proposed that S. epidermidis acts as a probiotic and prevents the colonization of pathogens such as S. aureus (2, 3). Staphylococcus epidermidis causes chronic infections associated with medical devices, such as intravascular catheter or prosthetic joint infections, vascular graft infection, surgical site infection, central nervous system shunt infection, and cardiac device infection (4).
Fbe acts as an adhesin, which can facilitate the interaction with fibrinogen. The expression of fbe is poor in vitro and increased in the presence of serum. In the presence of anti-fbe antibody, adherence of S. epidermidis to fibrinogen was blocked and infection was attenuated in animal models (5, 6). The ica operon, includes icaA, icaB, icaC and icaD genes and encodes polysaccharide intercellular adhesin (PIA), a microbial surface component-recognizing adhesive matrix molecule (MSCRAMM) of S. epidermidis that affects biofilm formation and is crucial for bacterial adhesion (7). The IS256 is a mobile genetic element present in the genome of S. epidermidis at several copy numbers. It is believed that IS256 is associated with virulence determinant genes, contributes to genetic adaptation, and is active in invasive strains during infection. Expression of ica operon may be affected by IS256. IS256 increases the production of PIA, which plays an important role in biofilm formation and immune system invasion (4, 8).
Accumulation-associated protein (aap) is a 140-KD protein essential for S. epidermidis accumulation on the surfaces by formation of Aap-based fibril-like structures, which can potentially underlie the formation of biofilm (9, 10). Biofilm-associated protein (Bap), a 239-KD surface protein, is found only in some S. aureus strains. Its equivalent protein in S. epidermidis is called Bap homologue protein (Bhp). Bap homologue protein plays an important role in bacterial adhesion and colonization and catalyzes biofilm formation (11).
2. Objectives
The aim of this study was to determine the prevalence of icaA, IS256, aap, fbe and bhp in clinical isolates of S. epidermidis.
3. Materials and Methods
3.1. Bacterial Isolates
This study had a cross-sectional design. Overall, 59 S. epidermidis strains, isolated from blood (n = 50), urine (n = 4), tracheal (n = 4) and wound (n = 1) samples, were obtained from patients at intensive care units of various hospitals of Tehran, Iran.
3.2. Staphylococcus epidermidis Identification
For S. epidermidis identification, the sample was cultured on sheep blood agar (Merck, Germany), overnight under aerobic conditions at 37°C; Staphylococci should produce white, smooth, medium colonies on sheep blood agar. Gram stain was performed for suspected colonies, after gram-positive cocci observation. Catalase test was performed and catalase positive colonies were cultured on mannitol salt agar (Merck, Germany), overnight under aerobic conditions at 37°C. Staphylococci can growth on mannitol salt agar while S. epidermidis and some other Staphylococci are not able to ferment mannitol, thus the medium remained red. Tube coagulase test with rabbit plasma was performed for mannitol negative colonies. Staphylococcus epidermidis and some other Staphylococci are coagulase negative and could not produce clot in the tube. Final identification of S. epidermidis was performed by the ability of urease production and carbohydrate fermentation. Staphylococcus epidermidis are able to produce urease, thus the medium turned red. It is also unable to ferment D-trehalose, thus the medium turned red, while it is able to ferment D-manose and D-maltose, thus the medium turned yellow (12, 13).
3.3. DNA Extraction
DNA of each S. epidermidis isolate was extracted from 1 mL of overnight bacterial culture. Extraction was performed by adding TE [10 mM Tris, 1 mM EDTA (pH = 8)] buffer and lysostaphin, boiling at 95°C for five minutes, then cooling at 0°C for five minutes, followed by centrifugation at 10000 rpm for five minutes. The supernatant was used as the template DNA in PCR reactions. DNA was measured using a BioPhotometer (Eppendorf, Germany) to determine the concentration and purity.
3.4. Detection of Virulence Determinants
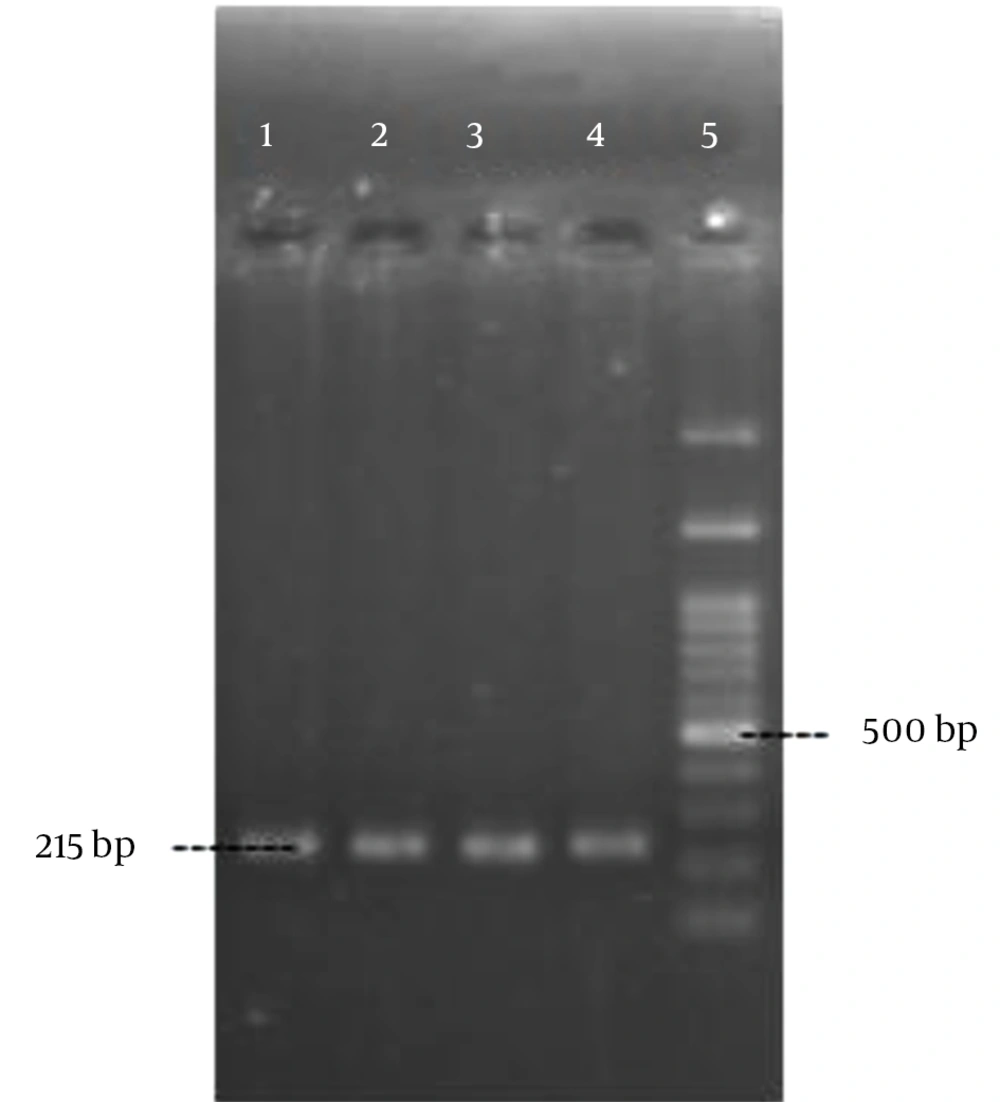
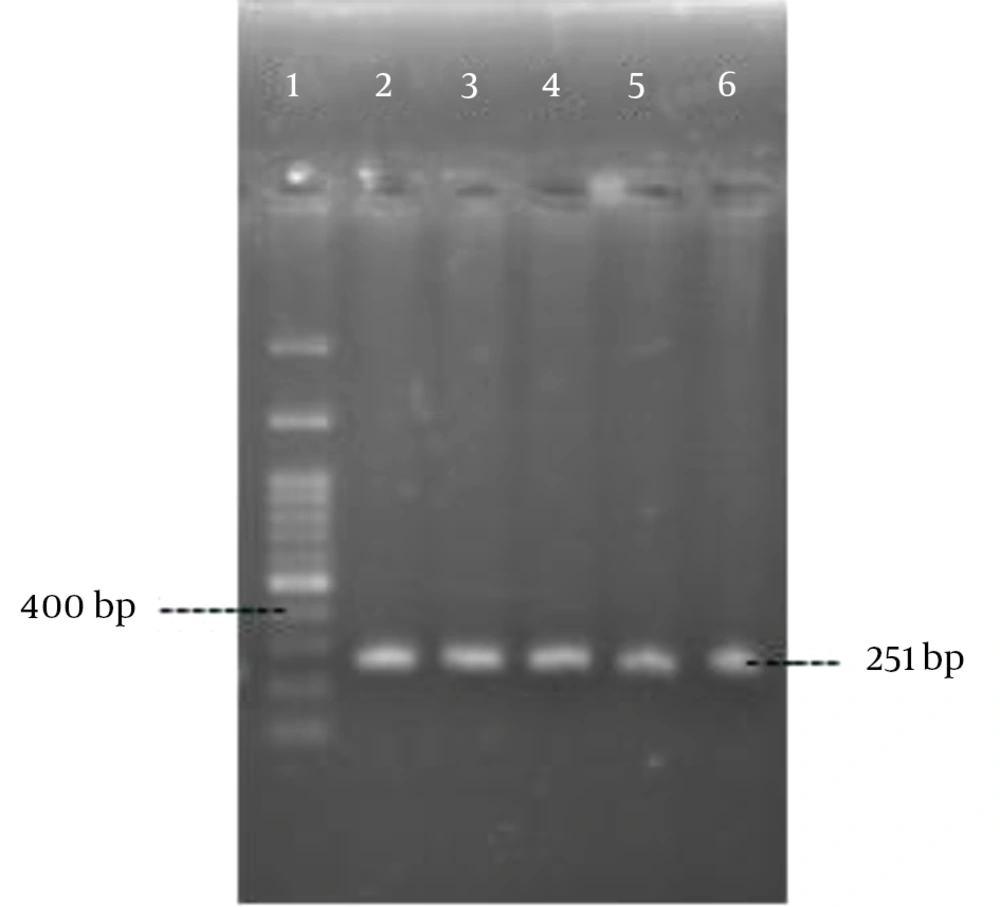
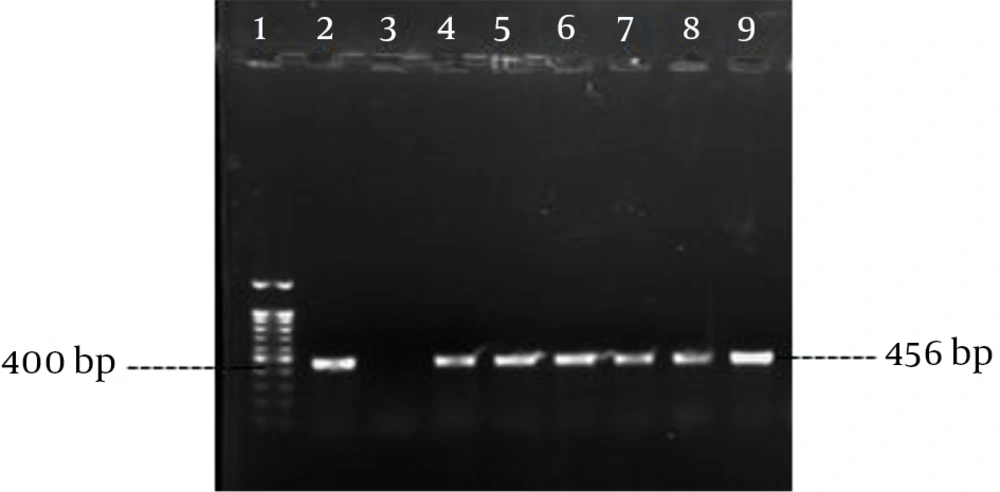
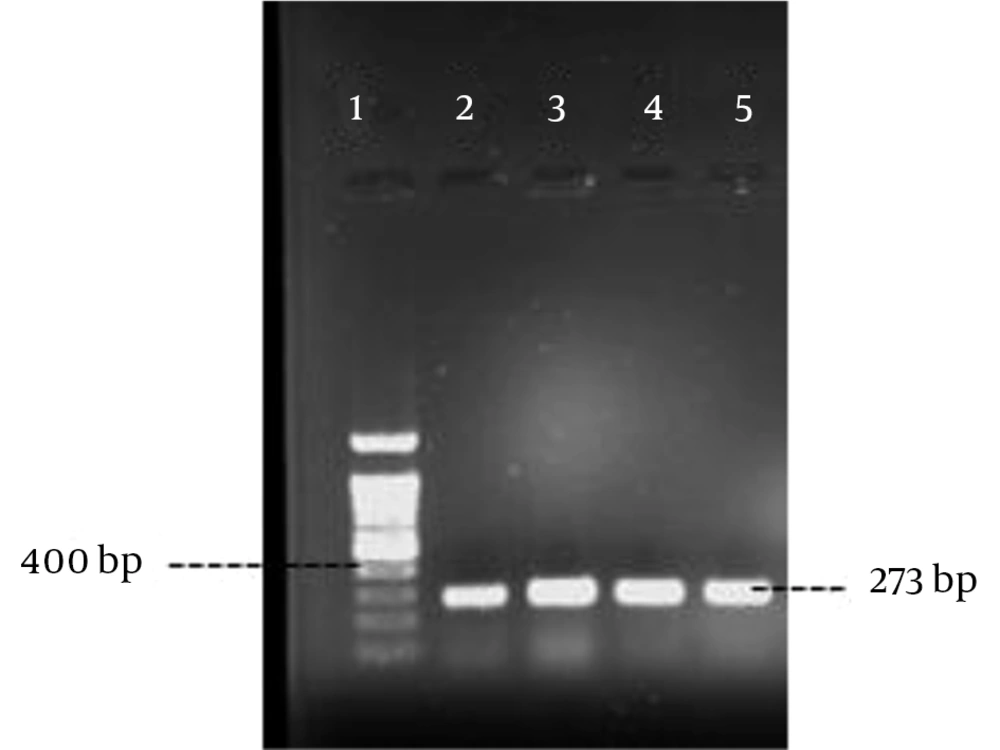
Polymerase chain reaction was performed as described by Rohde et al. (14) with the specific primers (Pishgam biotech, Iran) shown in Table 1. Amplification was performed in a MJ mini Gradient thermal cycler PTC-1148, USA. The amplified products were visualized by UV light after electrophoresis on 1% agarose gel (Figures 1 - 5). A positive control and negative control (reaction mixture without DNA) were included in each PCR run.
| Gene | Primer (5' → 3') | PCR Condition | Expected Size, bp |
|---|---|---|---|
| icaA | 5 min, 95°C; 30 cycles of: 30 s, 94°C; 30 s, 52°C; 50 s, 72°C; final extension, 4 min, 72°C | 215 | |
| Forward | TCGATGCGATTTGTTCAAACAT | ||
| Reverse | CTGTTTCATGGAAACTC | ||
| IS256 | 5 min, 95°C; 30 cycles of: 30 s, 94°C; 30 s, 52°C; 50 s, 72°C; final extension, 4 min, 72°C | 251 | |
| Forward | CCAGTTCATTTGGGTTTATAGC | ||
| Reverse | CATATACATGGCAAGCTCTAGG | ||
| fbe | 5 min, 95°C; 30 cycles of: 30 s, 94°C; 30 s, 60°C; 50 s, 72°C; final extension, 4 min, 72°C | 273 | |
| Forward | CTACAAGTTCAGGTCAAGGACAAGG | ||
| Reverse | GCGTCGGCGTATATCCTTCAG | ||
| aap | 5 min, 95°C; 30 cycles of: 30 s, 94°C; 30 s, 60°C; 50 s, 72°C; final extension, 4 min, 72°C | 465 | |
| Forward | AAACGGTGGTATCTTACGTGAA | ||
| Reverse | CAATGTTGCACCATCTAAATCAGCT | ||
| bhp | 5 min, 95°C; 30 cycles of: 30 s, 94°C; 30 s, 61°C; 90 s, 72°C; final extension, 4 min, 72°C | 1585 | |
| Forward | ATGGTATTAGCAAGCTCTCAGCTGG | ||
| Reverse | AGGGTTTCCATCTGGATCCG |
3.5. Statistical Analysis
Confidence interval test was used to assess the statistical significance with confidence level of 95% (α = 0.05).
4. Results
The most frequent virulence determinant in this study was fbe [89.8% (95% CI: 97.5 - 82.1)] and the least frequent virulence determinant was bhp [15.3% (95% CI: 24.5 – 6.1)]. The frequency of IS256, icaA and aap were 72.9% (95% CI: 84.2 – 61.6), 55.9% (95% CI: 68.6 – 43.3) and 64.4% (95% CI: 73.4 – 55.4), respectively. Coexistence of icaA and IS256 was observed in 44.1% (95%CI: 56.8 – 31.4) of the isolates. Coexistence of aap and icaA was seen in 39% (95% CI: 51.4 – 26.6) of the isolates. In six isolates [10.1% (95% CI: 17.8 – 2.4) only one of the investigated virulence determinants was observed, while in three isolates [5% (95% CI: 10.5 – 0)] all of the investigated virulence determinant was observed.
5. Discussion
Staphylococcus epidermidis, which was previously regarded only as part of the human flora, is now considered as an infectious agent that causes infections associated with medical devices. These infections may be transmitted from the patient’s skin to the hospital staff during device insertion. The adhesion ability of S. epidermidis is one of the most important mechanisms through which it causes infections (4, 15). Polysaccharide intercellular adhesin, which is produced by ica operon, catalyzes cell–cell adhesion and biofilm formation (16). Thus, ica operon, producing PIA and adhesion to host protein, coated medical devices are the two main determinants of S. epidermidis invasiveness (8, 17). Fbe, an adhesion protein present in S. epidermidis and S. aureus, facilitates binding to fibrinogen-coated device surfaces, which is an essential mechanism for bacterial persistence in or on the human body that leads to infections (18). Thus, ica operon and IS256 are the two determinants of S. epidermidis invasiveness, which play an important role in biofilm formation and immune system invasion (4, 8). It is believed that aap is one of the most important factors for PIA-independent biofilm formation in S. epidermidis (9). The increased expression of aap may lead to biofilm formation (19, 20). Furthermore, Bhp plays an important role in bacterial accumulation and adhesion and catalyzes biofilm formation (11, 21).
In our study, frequency of icaA was found to be 55.9%. Diemond-Hernandez et al. (22) reported that the frequency of icaA was 51.1% while Koskela et al. (8) reported that this frequency was 50%. Our results are not significantly different from those of these studies. Frequency of icaA was reported as 73% by Okee et al. (23), 88.6% by Gad et al. (16), and 88.6% by Rohde et al. (14). The frequency observed in our study was significantly lower than that reported in these studies. In our study, frequency of IS256 was found to be 72.9%, which is not significantly different from 81% reported by Koskela et al. (8); however, it is significantly lower than 93.8% reported by Rohde et al. (14) and 100% reported by Okee et al. (23) and significantly more than 61.1% reported by Mekni et al. (24). Fbe was found in 89.8% of the isolates, which is significantly less than 100% reported by Rohde et al. (14), and does not show significant difference with 50.8% reported by Mekni et al. (24). Overall, 15.3% of investigated isolates in this study harbored bhp which is not significantly different from 10% reported by Rohde et al. (14), 11.1% reported by Okee et al. (23), and 18.8 reported by Mekni et al. (24). In our study, the frequency of aap was found to be 64.4%, which is significantly higher than 17% reported by Okee et al. (23) yet significantly lower than 93.8% reported by Rohde et al. (14) and Pourmand et al. (19). Coexistence of aap and icaA was detected in 39% of the isolates in our study, which is not significantly different from 37.1% reported by Liduma et al. (25). Coexistence of IS256 and icaA was detected in 44.1% of the isolates in our study, which is significantly lower than 58% reported by Koskela et al. (8).
In conclusion, prevalence of fbe and aap observed in our study was significantly different from that reported by other similar studies; however, the frequency of bhp was in accordance with that reported by other studies. Prevalence of icaA and IS256 was not significantly different from that reported by some studies yet was significantly different from other studies.