1. Background
Biofilm-forming bacteria are often seen on the surfaces of tissues (1-3) and biomaterials at sites of persistent infection (4, 5). Medical implants and catheters are particularly susceptible to biofilm formation because immune responses are significantly reduced in proximity to foreign bodies (6, 7). In fact, biofilm formation is a main cause of implant failure and often limits the lifetime of many indwelling medical devices (8). Biofilms protect microorganisms from opsonization by antibodies, phagocytosis, and removal via the ciliary action of epithelial cells (5). In addition, bacterial populations in biofilms are considerably more resistant than free-living planktonic cells are to antibacterial agents (9, 10). Therefore, treatment of an infection after a biofilm has been established is frequently futile with the current therapy options (11).
Klebsiella pneumoniae is frequently found in a variety of environmental niches, such as soil, vegetation, and water, including the drinking-water distribution system. It is also an important nosocomial pathogen involved in urinary tract infections, hospital-acquired pneumonia (HAP), ventilator-associated pneumonia (VAP), surgical-wound infection, bacteremia, and septicemia (12-14). Interestingly, previous studies reported a high frequency of opportunistic infections caused by K. pneumoniae among persons with certain debilitating medical conditions, such as diabetes mellitus or bladder neuropathy (15, 16).
The most important virulence factors contributing to K. pneumoniae pathogenesis with regard to the severity of its infections are capsular polysaccharides, type 1 and type 3 pili, which can contribute to biofilm formation (14, 17, 18). The first biofilm-forming K. pneumoniae strain was described at the end of the 1988s (19). These bacteria can produce a thick layer of extracellular biofilm as a virulence factor that helps the organism attach to living or abiotic surfaces, preventing the effects of antimicrobial agents (20). For these reasons, detailed knowledge about biofilm formation and biofilm-producing bacteria is necessary for the management and treatment of related infections. In addition, the determination of genetic diversity for dominant species identification can be useful for preventing the spread of infection in hospitals. Knowing the dominant genotype of isolates can be helpful for the identification of the source of an infection, and for implementing preventive procedures and applying infection control.
2. Objectives
The present study was designed to evaluate biofilm development among K. pneumoniae clinical isolates, and to perform molecular genotyping using the enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) method.
3. Patients and Methods
3.1. Bacterial Isolation and Identification
In the present study, 94 K. pneumoniae strains were isolated at two hospitals in Tehran from February to July 2014. The K. pneumoniae strains were recovered from blood, sputum, surgical wound swabs, and urine samples of hospitalized patients. Definitive identification of isolates was performed using conventional biochemical tests (21). After recognition, the K. pneumoniae strains were stored in tryptic soy broth (TSB) (Merck Co., Germany) with 10% glycerol at -70°C. All isolates were freshly subcultured on brain-heart infusion (BHI) agar (Merck Co., Germany) prior to every analysis.
3.2. Biofilm Model
Fresh colonies of K. pneumoniae isolates were used to inoculate 5 mL of brain-heart infusion (BHI) broth medium (Merck Co., Germany). The cultures were incubated for 18 hours at 37°C with shaking at 200 rpm. Following incubation, the amount of cells in each culture was quantified and adjusted to 0.5 McFarland (1.5 × 108 CFU/ml), and 200 μl of each bacterial suspension was transported into eight wells of a 96-well microtiter plate containing BHI broth. Staphylococcus epidermidis RP62A and Pseudomonas aeruginosa PAO1, both of which are well-recognized as biofilm-forming strains, were used as two positive controls in the biofilm assays. Sterile BHI broth was incorporated as a negative control. A 96-peg plate was then located in the wells of the microtiter plate, allowing the pegs to be submerged within the bacterial culture. The inoculated peg plate was relocated to a 96-well microtiter plate containing fresh BHI broth. The inoculated microtiter plates were incubated on a shaking platform at 37°C for two days, permitting mature biofilms to be established. Each biofilm assessment was repeated on two distinct occasions.
3.3. Semi-quantification of Biofilm Biomass
The biofilm biomass assay was semi-quantified after the modification of a procedure described by Mowat et al. (22). After incubation, the peg plate was removed from the microtiter plate, then washed with PBS to remove planktonic free-floating cells that were weakly attached to the surface. The cells were dried for 30 min at 37°C. Each replicate peg was stained with 0.5% (w/v) crystal violet for 5 min, and surplus crystal violet was eliminated by carefully rinsing the peg plate under running distilled water. The stained biofilms were decolorized by the addition of 100 ml of 95% ethanol to each well for 1 min. The ethanol was relocated to a clean 96-well microtiter plate and the optical density (OD) was measured using a microtiter plate reader.
The biofilm-forming capacity of each test isolate was compared with the positive and negative controls by analyzing the absorbance of the crystal violet stain obtained for each biofilm. The strains were classified into four groups: non-adherent (< 25% absorbance compression with positive control), weakly adherent (25% - 50%), moderately adherent (51% - 75%), or highly adherent (76% - 100%), depending on the OD of the bacterial biofilm (Table 1).
| Biofilm Production, % | Classification |
|---|---|
| < 25 | None |
| 25 - 50 | Weak |
| 50 - 75 | Moderate |
| > 75 | High |
Classification of Biofilm-formation Capacity in Comparison With Positive Controls
3.4. Molecular Genotyping
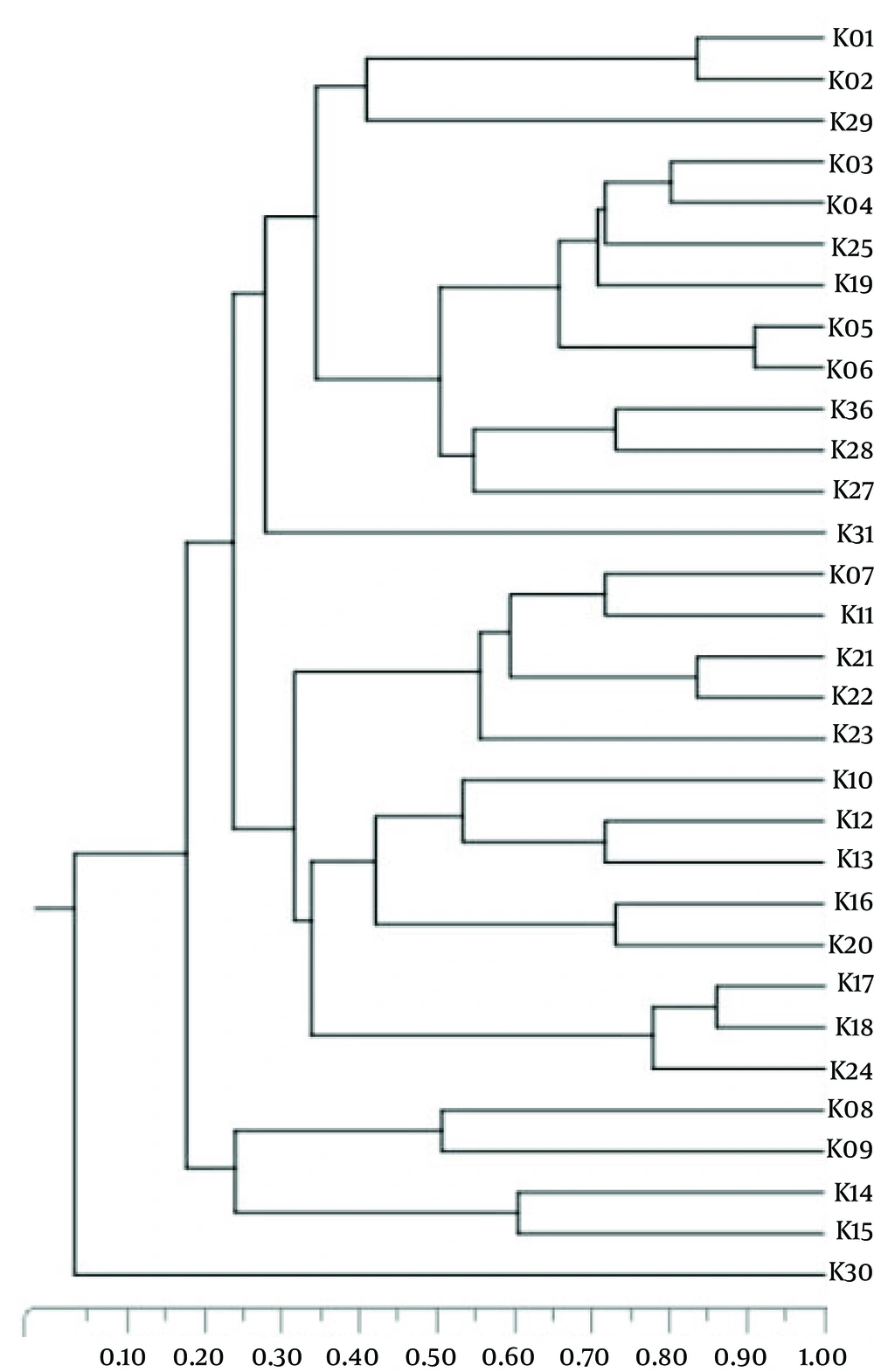
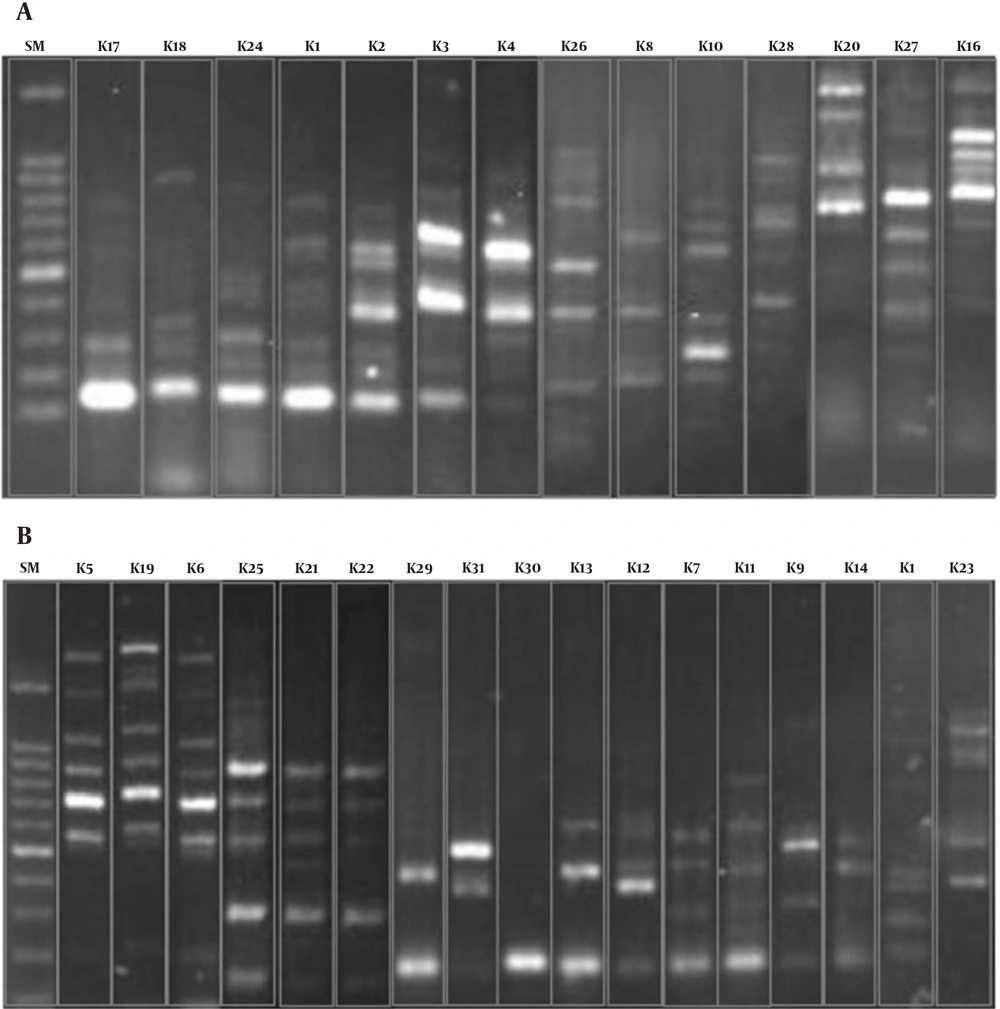
To evaluate the genetic relatedness among the isolates, the ERIC-PCR method was performed as described previously (23). The banding profiles of the isolates were included in different types by visual inspection. An isolate was considered a different type if its banding pattern differed by two or more bands. In other words, isolates were classified as identical types when their banding patterns were different in one band only (24). The K. pneumoniae genotypes were named by K1 to Kn. To determine the similarity rate among the obtained types, they were analyzed using the unweighted pair-group method with arithmetic mean (UPGMA).
3.5. Statistical Analysis
SPSS version 22.0 was used for statistical analysis. Chi-square analysis was used for comparisons between the capacity of biofilm production and the type of clinical specimen.
4. Results
4.1. Distribution of Samples
Of 94 K. pneumoniae clinical isolates, 58 (61.7%) were obtained from urine specimens, 17 (18.1%) from surgical wounds, 11 (11.7%), from sputum, and 8 (8.5%) from blood.
4.2. Biofilm Formation
Our findings indicated that 93.6% of K. pneumoniae isolates formed biofilms. The strains were divided into four categories as described above. According to the biofilm analysis, 52.1% (n = 49) of the 94 tested K. pneumoniae isolates were categorized as moderately biofilm-producing strains, 33% (n = 31) formed fully established biofilms, 8.5% (n = 8) produced negligible biofilms, and 6.4% (n = 6) of the strains were non-biofilm producers. The K. pneumoniae strains that were isolated from sputum, surgical-wound swabs, and urine samples had a remarkably greater ability to form fully and moderately established biofilms, compared with isolates obtained from blood samples (Table 2). high (+++) biofilm-formation ability among the isolates obtained from sputum was more than other isolates (P < 0.001) significantly.
| Biofilm Formation | Type of Specimen | ||||
|---|---|---|---|---|---|
| Blood | Sputum | Surgical-Wound Swab | Urine | Total | |
| None (0) | 4 (50) | 0 (0) | 0 (0) | 2 (3.5) | 6 (6.4) |
| Weak (+) | 3 (37.5) | 0 (0) | 1 (5.9) | 4 (6.9) | 8 (8.5) |
| Moderate (++) | 1 (12.5) | 3 (27.3) | 5 (29.4) | 40 (68.9) | 49 (52.1) |
| High (+++) | 0 (0) | 8 (72.7) | 11 (64.7) | 12 (20.7) | 31 (33) |
| Total | 8 (100) | 11 (100) | 17 (100) | 58 (100) | 94 (100) |
Correlation Between Type of Specimen and Biofilm Formationa
4.3. ERIC-PCR Typing
Genotyping of K. pneumoniae revealed 31 different ERIC types,while 11 isolates were non-typeable because they had no banding pattern after PCR reaction and electrophoresis. The K1 type was the most common and included 26 isolates, followed by type K2,which comprised 19 patterns. The K09 to K31 types were unique, and each contained one isolate exclusively. Figure 1 indicates the rates of genetic relatedness between the obtained ERIC types. The banding patterns of 31 types are shown in Figure 2.
5. Discussion
In the present study, we assayed the capacity for biofilm production in isolates of K. pneumoniae obtained from clinical specimens. Our findings indicated that an enormous proportion (more than 93%) of isolates were biofilm-producing strains. Biofilm-forming bacteria cause infections, such as catheter- and implant-related infections, as well as life-threatening disorders in patients with cystic fibrosis, chronic wounds, and chronic otitis media. These bacteria affect millions of people around the world every year, with a high mortality rate (25). The ability of nosocomial opportunistic microorganisms such as K. pneumoniae to produce biofilms on host-tissue surfaces is a critical stage in the development of infection. Biofilm formation affects the efficacy of antimicrobial therapies and the outcomes of subsequent infections (18, 26).
Yang and colleagues conducted a study in 2008 on biofilm formation by K. pneumoniae strains isolated from urine samples, sputum, wound swabs, and blood (27). They found that 62.5% of the isolates generated biofilms, which is less than the results obtained in our study. This may be due to differences in geographical area and sample size. Saeed and colleagues evaluated biofilm formation in three K. pneumoniae strains, and showed that all of them produced biofilms (28). The reason for the discrepancy in our results compared to their findings may be due to their small sample size (n = 3). Some studies, such as one by Saxena et al., evaluated biofilm production in other bacteria by assessing biofilm formation in P. aeruginosa with the qualitative tube method (29). The findings revealed that 20%, 21.25% and 58.75% of the isolates, respectively, were highly potent biofilm producers, moderate producers, or weak/non-biofilm producers. According to these results, the ability of K. pneumoniae strains to form biofilms may be higher than that of P. aeruginosa strains.
Clinical observations and experimental investigations have clearly confirmed that in most cases, antibiotic therapy alone is inadequate to eradicate biofilm-forming infections (30). Consequently, the successful treatment of biofilm-associated infections with currently accessible antibiotics, and the evaluation of outcomes, has become important and urgent for clinicians. PCR-based fingerprinting techniques, such as ERIC-PCR, are quick methods for typing the strains of K. pneumoniae. The ERIC-PCR method is most reliable when two or more band differences are used as the cutoff (24). In the present study, this method was used for molecular typing of K. pneumoniae isolates from two hospitals in Tehran. Most of the type K1 strains (20 out of 26) were isolated from urinary infections at one hospital, where the most prevalent nosocomial type was K1. Strong genetic similarity was seen between the K5 and K6 types, which were both obtained from the same hospital. The highly heterogeneous strains were most often isolated from urine, followed by wounds.
Cartelle et al. indicated that ERIC-PCR has a discriminatory power similar to that of pulsed-field gel electrophoresis (PFGE). Therefore, itis an acceptable technique for molecular classification compared to PFGE, as a gold standard method (24). In a previous study performed on various clinical samples, a high level of genetic diversity among K. pneumoniae strains was reported, which is similar to the results of the present study (31). This heterogeneity may be due to the survival of K. pneumoniae on dry surfaces in the hospital environment, making accidental contamination of patients possible.
In our study, we did not observe any correlation between ERIC type and biofilm formation; in other words, the ability of biofilm formation in a specific ERIC type was not seen. Our results were in contrast with those of Diago-Navarro et al., who showed that the K. pneumoniae isolates with a strong capacity for biofilm formation were classified in a particular sequence type (32). This contradiction may be related to differences in typing methods. Therefore, according to Diago-Navarro et al.’s study, multilocus sequence typing can be helpful in the determination of the dominant types of biofilm-producer isolates. Biofilm formation is an important cause of bacterial resistance to the immune system and to antibacterial agents (5, 9). New approaches, such as photodynamic therapy and the utilization of medicinal plants, have recently been introduced against bacterial biofilms (33, 34). We suggest the use of these strategies to eliminate biofilm-producing bacteria from the hospital environment and to treat infections.
In conclusion, our results showed that an enormous proportion of K. pneumoniae isolates from sputum and surgical-wound swabs produced fully established biofilms. In contrast, K. pneumoniae strains isolated from blood exhibited weaker tendencies to form massive biofilms. Given these findings, there is reason to assume a relationship between infection and biofilm formation. In molecular genotyping, no correlation between ERIC types and biofilm-formation ability was observed. Therefore, other molecular genotyping methods are useful. Supplementary research into the mechanisms of biofilm formation in K. pneumoniae, as well as the design and use of new and effective approaches, will ultimately aid in the treatment of biofilm-mediated infections and in the reduction of morbidity and mortality in patients suffering from life-threatening nosocomial infections.