1. Background
Helicobacter pylori infection in humans is associated with the development of numerous gastric pathologies such as peptic ulcer, chronic gastritis and gastric cancer. Once established, the infected individuals carry H. pylori in their stomach yet if untreated with antibiotics, they can persist in the body for a lifetime. During the last decade, the rate of multidrug resistance among clinical H. pylori isolates has increased, which could be a result of widespread and non-appropriate use of various antibiotics (1, 2). By evaluation of resistance to clarithromycin, metronidazole, quinolones, amoxicillin and tetracycline, the investigators found that point mutations, redox intracellular potential, efflux pump systems and membrane permeability were the most common mechanisms of resistance in H. pylori (3).
Among the mechanisms involved in multidrug-resistance, active pumping out of the drugs by efflux pumps may be an important mechanism by which the gram negative bacteria may be protected from toxic effects of antibiotics (4-6). In general, the efflux pumps of gram-negative bacteria consist of an inner-membrane pump, a periplasmic adaptor protein and an outer-membrane channel in which the first one (inner membrane protein) acts with the second (periplasmic protein) and third (outer membrane protein). These efficiently structured complexes can cause the multidrug-resistance phenotype, which provides an efficient means for the export of structurally unrelated drugs (7, 8).
By analyzing H. pylori 26695 genome, 27 putative translocase have been identified that include ATP-Binding cassette (ABC) super family, major facilitator super family (MFS), resistance-nodulation-cell division (RND) family and multidrug and toxic compound extrusion (MATE) proteins (9). One of such translocases, which is found in H. pylori 26695 genome is hp1184 that belong to the MATE family (10). As proposed by Van Amsterdam et al., and Johnson et al., mutation in hp1184 can result in the susceptibility of H. pylori to EtBr (8, 9). hp1181 is another putative translocase, which may be related to the major facilitator super family. Analysis of hp1181 gene product in H. pylori has revealed that hp1181 could be an integral membrane protein, which can contribute to resistance via a multi-drug resistance efflux protein (11, 12).
2. Objectives
The results of each basic study on the mechanisms of multidrug resistance in the case of H. pylori may be beneficial for designing more suitable antibiotic combinations for successful treatment of infection. The aim of the present study was to evaluate the association between presence of two predicted efflux-related genes, hp1181 and hp1184, in MDR clinical strains of H. pylori and their role in the active-efflux of antibiotics.
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
The MDR strains employed in this study were selected from screening of the H. pylori strains isolated by culturing of biopsy specimens from children admitted to the medical center of Tehran for peptic disease problem (13). The isolated strains were plated onto modified Campy blood agar containing brucella agar base (Merck, Germany), supplemented with 5% defibrinated sheep blood and antibiotics. The plates were incubated at 37°C under microaerophilic atmosphere for three to seven days. The grown colonies were identified by Gram staining, standard biochemical tests and PCR using H. pylori specific primers for 16SrRNA and ureC (14, 15). Helicobacter pylori strain 26695 (from CNRS, Bordeaux, France) for which complete sequence and predicted proteins are available, was used as a positive control for presence of genes studied in PCR analysis (12). ATCC43504 strains (from CNRS, Bordeaux, France) was the reference strain for quality control of antibiotic susceptibility testing (16).
3.2. Determination of Minimum Inhibitory Concentration for Ethidium Bromide
Determination of minimal inhibitory concentration (MIC) for Ethidium Bromide (EtBr) was performed by the disc diffusion method. For this purpose, three-day-old bacterial cultures with a Mc-Farland opacity of 2.0 were cultured on Muller Hinton agar (Merck, Germany) plates supplemented with 7% fresh sheep blood. Plates were dried for five minutes, and the blank discs containing EtBr (Merck, Germany), with concentration of 4 to 64 μg/mL were placed on their surfaces; the inhibition zones of resistant strains were compared with the susceptible ones (13). The cut off for MIC determination of EtBr was considered 4 μg/mL.
3.3. Selection of Multi-Drug Resistant Strains
To select the MDR-strains, H. pylori isolates were screened for resistance to Amoxicillin (AMX), Ampicillin (AMP), Ciprofloxacin (CIP), Erythromycin (ERY) (Zakaria and Exir, Iran), Metronidazole (MTZ), and Tetracycline (TET) (Sigma, USA), using the minimal inhibitory concentration method. For this purpose, plates were prepared with Mueller Hinton (Merck, Germany) agar plus 7% fresh sheep blood and an appropriate concentration of antibiotics from 0.5 to 64 μg/mL. The plates containing antibiotics were then spread with bacterial suspension and incubated for two to three days at 37°C under microaerobic conditions. The cut off values for MIC determination was 4, 4, 4, 4, 8 and 8 μg/mL for AMP, AMX, TET, CIP, MTZ and ERY, respectively, according to the breakpoints previously described (13, 16). The MIC for all strains was compared in the presence and absence of CCCP, as previously described (13, 16, 17).
3.4. Measurement of Antibiotics and EtBr Accumulation
Accumulation assays were performed according to a previously described protocol with some modifications (13). Briefly, H. pylori strains were grown on campy blood agar for two days harvested and resuspended in pre-warmed brucella broth (37°C) to an optical density of 0.5. The cell suspension was incubated for 10 minutes at 37°C, and then centrifuged, washed and resuspended in 15 mM sodium phosphate buffer (pH 7.2). Uptake assays were initiated by the addition of an antibiotic or EtBr to the bacterial cell suspension at a final concentration of 10 µg/mL. Eight minutes after antibiotic or EtBr addition, CCCP was added to one-half of the reaction mixture at a final concentration of 200 µM and the other half was used as the control. After 15 minutes, each of the collected samples were placed at 4°C for one minute (to stop the function of the efflux pumps), centrifuged at 6000 g for 15 minutes, and the supernatants were used to measure the fluorescence of the respective antibiotics with a Shimadzu RF 5000 spectrofluorometer (Shimadzu scientific instruments) at excitation and emission wavelengths of 400 and 450 for AMP, AMX and TET; 300 and 350 for ERT; 279 and 447 for CIP; and 544 and 590 for EtBr, respectively. These wavelengths have previously been selected by testing various experimental conditions (13).
The pellet was used for measurement of the dry weight of bacteria by subtracting the initial weight of the cell-containing vial. Concentration of antibiotics in the supernatants were calculated using a standard curve of the respective antibiotics (concentration ranging from 100 to 1000 ng) in 0.1 M of glycine hydrochloride pH 3.0 .The results were expressed as ng of antibiotics and EtBr per mg (dry wt) of bacteria.
3.5. Polymerase Chain Reaction
DNA was extracted using the phenol-chloroform method, as previously described (14). Based on the sequence alignments of the genes from H. pylori 26695 strain in gene bank, specific hp1181 primers were designed as follow: F, 5'AAG AAG AAG AGC GCA CCAA3' and R, 5'GCT CTA AGG TGG CAA AAC C3'. These PCR primers were predicted to amplify a 579-bp amplicon. The reaction consisted of 32 cycles, with each cycle composed of one minutes at 95°C, three minutes at 60°C and four minutes at 72 °C. After a final extension of 15 minutes at 72°C, the amplicons were electrophoresed on 1.2% agarose gel. The gene specific hp1184 primers were prepared according to the primers described by Van Amsterdam as follow: F, 5'ACG CTC TAG ACT CAA AGC ACG GCA GAA TTT3'; R, 5'ACG CCT CGA GGA ACT TTT GGT GCG TTG GAT3', which predicted to amplify a 445-bp amplicon. The reaction consisted of 32 cycles, with each cycle composed of one minute at 95°C, four minutes at 59°C and seven minutes at 72°C. For each PCR reaction, a PCR control without template DNA was used. After a final extension of 15 minutes at 72°C, the amplicons were electrophoresed on 1.2% agarose gel (7).
3.6. Evaluation of hp1181 gene expression by RT-polymerase Chain Reaction
RT-PCR was performed to compare the difference in expression of hp1181 between 26695 susceptible (standard) strain and MDR clinical strains. For this purpose, the concentration of high-pure RNA, prepared according to the manufacturer’s recommendations (Roche, Germany), was measured at 260 nm and the cDNA was synthesized using first-strand cDNA synthesis kit for RT-PCR (Roche, Germany). The reaction for cDNA synthesis was held at 25°C for ten minutes, 42°C for 60 minutes and 99°C for five minutes, followed by PCR as described above. The RT-PCR-products were visualized by electrophoresis on 1% agarose gel and 15% acrylamide gel with a 100-bp ladder size-marker (Cinnagene, Iran). The RT-PCR analysis of hp1181 was performed for MDR strains, susceptible and 26695 H. pylori strains. The size of products for the 26695 susceptible strain was compared with that of MDR strains and was calculated by comparison with standard of MW. This reaction was repeated two times.
4. Results
Multi-resistance or resistance patterns of the strains, according to previously described representation (12), are shown in Table 1. These MDR strains have been classified into three groups, according to their behavior toward active efflux inhibition by CCCP. As demonstrated in Table 2, in the presence of CCCP, the MICs of eight MDR strains to EtBr was decreased by two to four folds; five of them also showed a two-fold reduction in their MIC for antibiotics (AMX, AMP, TET, CIP and MTZ). Accumulation assay for EtBr, exhibited active efflux for eight MDR strains (S3, S11, S13, S1, S2, S4, S9 and S12), of which five demonstrated active efflux for all of the antibiotics except ERY (Table 3).
In the presence of CCCP, the increase in the amounts of accumulated EtBr for the strains of group I and II was two folds and the increase in those of antibiotic for the strains of group II, was between 1.4 and 1.8 folds, which was lowest for β-lactam, and highest for TET (Table 3).
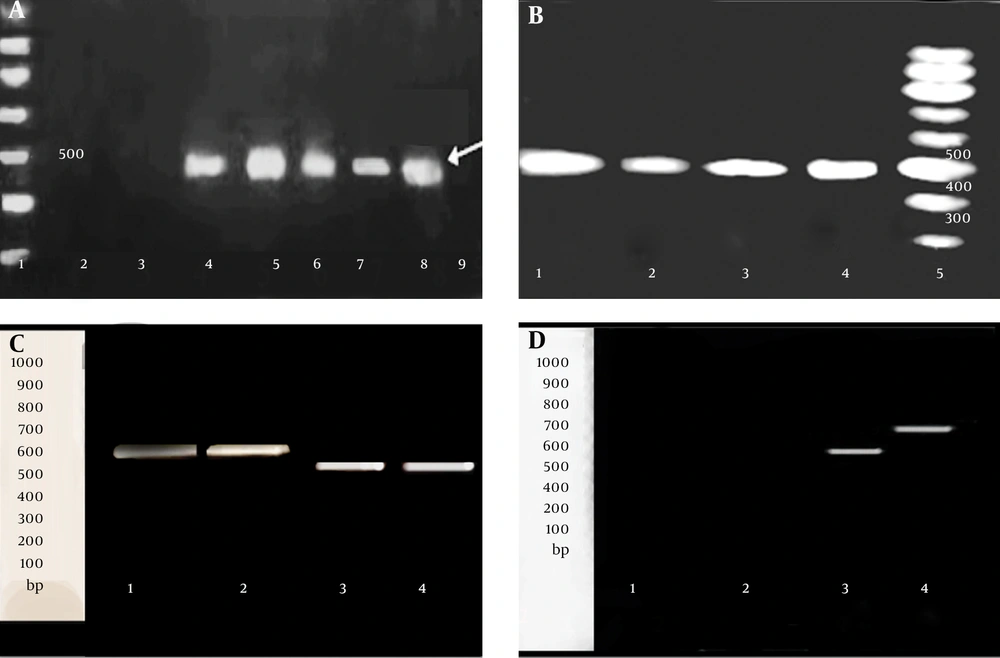
In the presence of CCCP, no difference in MIC for EtBr or antibiotics, as well as no difference in the amount of this group comprised of the following strains, S7, S10 and S13, which were only resistant to β-lactams and strains, S5, S6, S8 and S14, which were MDR. Two strains, S16 and S17, susceptible to all of the antibiotics and EtBr, demonstrating no efflux phenotype for them, amplified neither for hp1184, nor for hp1181. The relationship between the presence of active efflux and presence of two genes hp1181 and hp1184, are demonstrated in Table 2. The results of PCR for the detection of hp1184 (a product of 445 bp) and hp1181 (a product of 579 bp) are shown in Figure 1A - D. Standard 26695 and ATCC43504 strains, which were both susceptible to the entire antibiotic tested in this work, showed no efflux for EtBr or antibiotics. To understand the status of hp1181 gene in the MDR strains as well as in 26695 standard strain, they were analyzed by RT-PCR. The product of RT-PCR for hp1181 in the MDR strains was approximately 100 bp shorter than standard 26695 susceptible strain (Figure 1C - D).
| Resistance Patterns | Number of Strains | Designed Strain |
|---|---|---|
| β-lactame, TET, CIP, MTZ, ERY, EtBr | 3 | S1, S2, S12 |
| β-lactamas, TET, MTZ, ERY, EtBr | 3 | S4, S6, S8 |
| β-lactamas, TET, CIP, ERY, EtBr | 1 | S3 |
| β-lactamas, TET, CIP, EtBr | 2 | S9, S15 |
| β-lactamas, TET, ERY, EtBr | 2 | S11, S14 |
| β-lactamas, MTZ ,ERY, EtBr | 1 | S5 |
| β-lactamas | 3 | S7, S10, S13 |
| Susceptible to β-lactamas, TET, CIP, MTZ, ERY, EtBr | 2 | S16, S17 |
| Strains | AMP, -/+ | AMX, -/+ | TET, -/+ | CIP, -/+ | MTZ, -/+ | ERY, -/+ | EtBr, -/+ | hp1181 | hp1184 |
|---|---|---|---|---|---|---|---|---|---|
| Group I MDR: Decreased MIC for EtBr With CCCP | |||||||||
| S3 | 16/16 | 8/8 | 8/8 | 8/8 | 8/8 | 16/16 | 32/8 | - | + |
| S11 | 16/16 | 16/16 | 8/8 | 4/4 | 8/8 | 16/16 | 8/4 | - | + |
| S13 | 8/8 | 8/8 | 4/4 | 4/4 | 8/8 | 8/8 | 16/4 | - | + |
| Group II MDR: Decreased MIC for Both EtBr and Antibiotics With CCCP | |||||||||
| S1 | 16/8 | 16/8 | 8/4 | 8/4 | 16/8 | 16/16 | 32/8 | + | + |
| S2 | 8/4 | 8/4 | 8/4 | 8/4 | 16/8 | 16/16 | 16/8 | + | + |
| S4 | 8/4 | 8/4 | 8/4 | 4/2 | 16/4 | 16/16 | 8/4 | + | + |
| S9 | 16/8 | 16/8 | 8/4 | 8/4 | 8/4 | 8/8 | 16/4 | + | - |
| S12 | 16/8 | 16/8 | 8/4 | 8/4 | 16/8 | 16/16 | 16/8 | + | - |
| Group III MDR: No Difference in MIC With CCCP | |||||||||
| S5 | 8/8 | 8/8 | 4/4 | 4/4 | 16/16 | 16/16 | 8/8 | - | - |
| S6 | 16/16 | 16/16 | 8/8 | 4/4 | 16/16 | 16/16 | 4/4 | - | - |
| S7 | 8/8 | 8/8 | 4/4 | 4/4 | 4/4 | 8/8 | 8/8 | - | - |
| S8 | 8/8 | 8/8 | 8/8 | 2/2 | 16/16 | 16/16 | 8/8 | - | - |
| S10 | 8/8 | 8/8 | 4/4 | 2/2 | 8/8 | 8/8 | 4/4 | - | - |
| S14 | 8/8 | 8/8 | 8/8 | 4/4 | 8/8 | 16/16 | 8/8 | - | - |
| S15 | 16/16 | 16/16 | 8/8 | 8/8 | 8/8 | 8/8 | 16/16 | - | - |
| Group IV: Susceptible Strains | |||||||||
| S16 | 0.5 /ND | 0.5/ND | 1/ND | 2/ND | 2/ND | 2/ND | 2/ND | - | - |
| S17 | 0.5 /ND | 0.5 /ND | 1/ND | 2/ND | 4/ND | 2/ND | 2/ND | - | - |
| 26695 | 2/ND | 2/ND | 2/ND | 1/ND | 4/ND | 4/ND | 2/ND | + | + |
Abbreviation: ND, Not determined.
aCut off values for MIC determination was 4, 4, 4, 4, 8; 8 , and 4 μg/mL for AMP, AMX, TET, CIP, MTZ, ERY, and EtBr.
| Strain /Groupb | EtBr +/-CCCP | AMP +/-CCCP | AMX +/-CCCP | ERY +/-CCCP | CIP +/-CCCP | MET +/-CCCP | TET +/-CCCP |
|---|---|---|---|---|---|---|---|
| S1/II | 32.69/17.98 | 20.54/15.00 | 17.94/13.25 | 16.03/16.0 | 32.60/18.51 | 30.25/16.86 | 43.10/23.63 |
| S2/II | 62.12/25.42 | 18.69/14.21 | 19.08/14.65 | 10.60/10.55 | 36.29/19.25 | 30.69/16.50 | 42.58/22.01 |
| S3/I | 59.40/32.4 | 24.00/23.8 | 18.30/18.23 | 23.08/23.10 | 13.98/14.01 | 24.42/24.60 | 19.10/18.96 |
| S4/II | 58.19/26.59 | 24.10/16.74 | 21.82/15.48 | 18.18/18.16 | 31.02/17.28 | 36.12/21.41 | 34.59/17.65 |
| S5/III | 26.98/27.01 | 18.71/18.69 | 17.19/17.21 | 14.14/14.10 | 14.91/14.86 | 17.56/17.60 | 19/61/19.59 |
| S6/III | 22.00/22.01 | 17.05/16.98 | 17.00/17.03 | 15.88/15.86 | 20.00/20.03 | 21.14/21.12 | 18.25/18.20 |
| S7/III | 26.11/26.12 | 19.00/18.96 | 16.83/16.85 | 14.71/14.69 | 22.00/22.03 | 21.10/21.07 | 17.00/16.98 |
| S8/III | 17.91/17.89 | 18.14/18.12 | 16.00/16.01 | 13.14/13.12 | 22.85/22.86 | 19.63/19.59 | 16.18/16.20 |
| S9/II | 37.15/18.85 | 24.08/18.36 | 18.31/14.09 | 12.65/12.63 | 21.70/15.53 | 25.40/17.43 | 23.00/14.00 |
| S10/III | 15.91/15.88 | 13.44/13.40 | 14.06/14.0 | 21.56/21.54 | 19.83/19.80 | 17.61/17.58 | 20.90:20.86 |
| S11/I | 49.51/22.56 | 15.28/15.23 | 14.90/14.85 | 12.26/12.23 | 20.06/20.0 | 18.00/17.98 | 24.67/24.65 |
| S12/II | 47.85/22.81 | 24.18/17.53 | 18.58/14.0 | 14.10/14.08 | 22.58/16.0 | 25.32/17.58 | 23.00/14.56 |
| S13/I | 39.76/19.63 | 18.06/17.98 | 17.26/17.21 | 14.55/14.55 | 21.68/21.64 | 15.85/15.87 | 17.32/17.28 |
| S14/III | 21.85/21.80 | 17.30/17.25 | 19.00/18.96 | 12.08/12.05 | 23.09/23.06 | 16.91/16.87 | 18.27/18.25 |
| S15/III | 27.10/27.02 | 24.05/23.98 | 22.51/22.46 | 18.06/18.00 | 20.81/20.78 | 24.64/24.67 | 15.90/15.86 |
| S16/IV | 45.90/45.86 | 35.71/35.68 | 33.76/33.76 | 33.14/33.12 | 35.90/35.85 | 40.10/40.06 | 30.15/30.12 |
| S17/IV | 65.46/65.43 | 44.06/43.98 | 42.00/42.02 | 49.03/48.97 | 39.84/39.78 | 52.98/52.93 | 61.58/61.56 |
| St | 41.73/41.65 | 50.12/46.96 | 39.86/39.79 | 33.16/33.06 | 44.61/44.57 | 36.03/35.91 | 32.61/32.56 |
aValues are expressed as ng of antibiotics and EtBr per mg (dry wt) of bacteria.
bgroups I - IV are classified as the Table 2, St: strain 26695.
A, Detection of hp1181: 1: 100-bp ladder; 2: S17; 3: S16; 4: S12; 5: 26695 standard strain, 6: S4; 7: S2; 8: S1; 9, control (no template); B, detection of hp1184: 1: 26695 standard strain, 2: S3; 3: S2; 4: S1; 5: 100-bp ladder; C, RT-PCR detection of hp1181 gene on 1% agarose gel. At the left: 100-bp standard; 1-2: 26695 standard strain; 3-4: S12; D, RT-PCR detection of hp1181 gene on 15% polyacrylamide gel. At the left: 100-bp standard; 1: control negative for RT-PCR; 2: S16, 3: S12, 5: 26695 standard strain.
5. Discussion
We compared the relationship between resistance to non-related antibiotics and their active efflux by evaluation of antibiotic accumulation in the bacterial cells using CCCP as a protonophore. This compound reduces the transmembrane electrochemical gradient and inhibits the efflux pumps (5, 7, 16). Although in our previous work (13), we demonstrated the efflux properties of the strains used in this work, the differences of antibiotic accumulation in the presence and absence of CCCP, was higher with the optimized protocol used in the present work. We found that decrease in MIC to antibiotics, as well as decrease in MIC to EtBr, were associated with an increase in their accumulation (Table 2). There was an association between this phenotype and presence of hp1184 and or hp1181. However, observation of a positive hp1184 status in only six strains demonstrating efflux phenotype for only EtBr, suggested the more specific role of hp1184 in the active efflux of EtBr (Table 2). This finding is consistent with the results of Van Amsterdam et al. (9).
We observed that decreases in MIC to EtBr and antibiotics, associated with increase in their accumulation amounts, was associated with the presence of hp1181 (100%) and hp1184 (50%) among the strains of group II (Tables 2 - 3). This observation indicated the involvement of hP1181 and hp1184 in their active efflux-phenotype. However, only five out of 12 MDR-strains, which demonstrated active efflux phenotype for β-lactam, TET, MTZ and CIP, had a positive status for hp1181, suggesting the more specific role of hp1181 gene in the efflux of these antibiotics (9, 10). Nevertheless, despite resistance to CLA, no changes either in MIC or accumulation amount was observed in the presence or absence of CCCP. Using Phe-Arg-b-naphthylamide (PAbN), as an efflux pump inhibitor, Hirata et al., proposed the contribution of efflux pumps to CLA resistance in Helicobacter pylori (18). They suggested that the RND-type multidrug efflux pump was involved in CLA resistance. Therefore, the difference between our results and those of Hirata et al. is related to the nature of efflux pumps involved in the resistance to CLA; which is RND-type multidrug efflux pump for CLA and MATE for the hp1184 and hp1181 MDR efflux pumps (10). Furthermore, as RND family would be responsible for macrolide intrinsic resistance in H. pylori, the nature of its specific inhibitors would also be different since it was demonstrated that various inhibitors influence the efflux pump behaviors (19, 20).
Consistent with the results discussed above, an association was observed between the absence of active efflux either for EtBr or for antibiotics and absence of either hp1184 or hp1181 genes in the strains of group IV, as well as in the susceptible strains. Therefore, acquisition of multidrug resistance in MDR strains is due to their active efflux phenotypes and may be associated with presence of hp1181 and hp1184 genes. However, 26695 H. pylori strain, which was susceptible to all of the antibiotics, contained both of hp1181 and hp1184 genes. The results of RT-PCR performed for the MDR strains and for 26695 control H. pylori strain could explain this question. The RT-PCR for hp1181 was positive for both MDR strains containing hp1181, and for 26695 strain, however, the size of RT-PCR product in the MDR strain was approximately 100-bp smaller than that of the 26695 strain. The same was observed for hp1184 (data not shown). A similar result has been previously observed in the expression of hp1165 gene in tetracycline-resistant strains of H. pylori strains (21).
According to the hypothesis postulated in our objective, we could show the presence of an association between hp1184 gene and the active efflux of EtBr. Also, we demonstrated the association of hp1181 gene expression with the specific efflux of non-related antibiotics among clinical MDR strains of H. pylori.
In conclusion, expression of the genes hp1184 and hp1181 is associated with the active efflux of EtBr and non-related antibiotics, respectively. However, for displaying these phenotypes, a post-transcriptional regulation step may be required