1. Background
Cytomegalovirus (CMV) infection is a frequent viral complication that occurs early post-kidney transplantation, and has been associated with increased graft rejection, morbidity and mortality (1, 2). It latently persists in the kidney and reactivates under immunosuppressive conditions, and causes direct and indirect clinical effects post-transplantation (2, 3). The nature of the specific immunologic defects predisposing kidney transplant patients to CMV infection and disease has also been partially elucidated (3). CMV may have substantial impact on host immune responses. Following infection, CMV infiltrates the cell and produces immediate-early antigens that regulate DNA production. During the ensuing 6 to 24 hours, CMV produces late antigens that direct nucleocapsid protein production. It also causes up regulation of IL-2 (IL-2) and can prevent the inhibition of IL-2 gene production by cyclosporine (4). CMV also down regulates MHC-1 molecules on the surface of infected cells to evade host immune recognition (4). On the other hand, hepatitis C virus (HCV) is a well-known agent of acute and especially chronic liver diseases (5). HCV infection has also been established as an important factor in reducing graft and patient survival post-kidney transplantation (6-10).
Successful acceptance of the transplanted kidney requires proper balance in immune responses with minimal inflammatory and immunological allograft damages influenced by viral infections. Both antigen-specific and non-specific co-stimulatory signals are necessary for increasing the duration of graft surveillance and control of virus related clinical outcomes post-transplantation (11, 12). Therefore, in the past few years, study of the determinative role of T cell co-stimulatory activator and inhibitory pathways was focused in transplant immunology (13). The co-stimulatory molecules include the CD28 family, which contains three closely related proteins, CD28, cytotoxic T-lymphocyte associated protein 4 (CTLA4) and inducible T cell costimulator (ICOS), and also the more distantly related programmed cell death 1 (PDCD1) (14). Their ligands expressed on antigen presenting cells are members of the B7 family (15, 16). The encoding genes of CD28, CTLA4, ICOS and PDCD1 in human are localized on chromosome 2. The CD28, CTLA4 and ICOS genes are closely linked at 2q33.3 (17, 18). The CD28 and ICOS have been widely recognized as the major positive co-stimulation pathways for T cell activation and play a central role in immune responses against viruses and graft rejection (19). On the other hand, CTLA4 and PDCD1 play fundamental roles in controlling T cell reactivity to self-antigens via transmission of inhibitory signal to T cells. They have important roles in maintaining tolerance to self-antigens as a negative regulator of T cell activation in transplant patients (20).
Several polymorphisms take part in co-stimulatory molecules and affect regulation of activator and inhibitory functions of these proteins in transplant patients (21). Recent studies reported associations between co-stimulatory molecule gene polymorphisms with CMV- and HCV-related clinical outcomes in transplant patients (22, 23). Furthermore, CTLA4 gene polymorphisms have serious effects on a variety of clinical courses accompanied by CMV infection (23, 24). Over expression of PDCD-1 increased by gene polymorphisms in HCV and CMV specific CD8–T cells, has been shown to diminish their antiviral abilities (25-27).
Haplotype-tagging single nucleotide polymorphisms (SNPs) of CTLA4 at position +49 in exon 1 and in position -318 of promoter were examined in individuals treated for chronic HCV (28). The +49G allele alone and in a haplotype with -318C was associated with an increased frequency of sustained HCV response to alpha interferon and ribavirin (28, 29). Also, associations between co-stimulatory molecule gene polymorphisms with kidney graft surveillance were reported. One cohort study included 141 kidney recipients and 229 healthy subjects to examine the distribution of genotypes and allele frequencies of CTLA4 and CD28 genes (30, 31). In another study, 325 Korean kidney recipients were examined to define the distribution of genotypes and allele frequencies of CTLA4 gene polymorphisms associated with susceptibility to acute and late acute rejection episodes (32).
2. Objectives
Based on these earlier reports, the possible associations between CTLA4, PD-1, ICOS, and CD28 co-stimulatory molecule gene polymorphisms with viral infections were evaluated in kidney transplant patients.
3. Patients and Methods
3.1. Patients
In this study a total of 172 patients, who had undergone kidney transplantation at the transplant ward at Namazi hospital, affiliated to the Shiraz University of Medical Sciences, Shiraz, Iran, were consecutively enrolled between years 2006 and 2010. Patients were followed for the graft outcome and acute rejection episode(s) during at least six months post-transplantation. Transplant patients were divided into two studied groups according to the presence (AR group) or absence (non-AR control group) of acute rejection episodes. The ethics committee of Shiraz University of Medical Sciences approved the protocol, which conformed to the ethical guidelines of the 1975 Helsinki declaration, and a written informed consent was obtained from all subjects.
Donors were selected on the basis of ABO blood group compatibility and all the patients were negative for lymph cytotoxic cross matches. Rejection episodes were identified by an expert nephrology team, based on approved clinical diagnostic criteria, and invariability was confirmed by needle biopsy as well as elevated serum creatinine and blood urea nitrogen levels (33). The standard immunosuppressive regimen included cyclosporine (5 mg.kg initially, then a maintenance dose of 2 to 2.5 mg.kg; cyclosporine level was 50 to 150 ng/mL), prednisolone (120 mg/d initially, tapering to 10 mg/d), and mycophenolate mofetil (1000 mg twice daily). Acute rejection was initially treated with intravenous steroids and steroid-resistant rejection was treated with OKT3 monoclonal antibody.
3.2. Co-stimulatory Molecule Gene Polymorphisms
3.2.1. DNA Extraction Protocol
An EDTA-treated blood sample was collected from each kidney transplant patient and buffy coat and plasma were harvested from each of them and preserved at -80°C until use. Genomic DNA was extracted from buffy coats using a QIAamp DNA mini kit (Qiagen, Germany), according to the manufacturer’s instructions.
3.2.2. Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) Protocols
The SNPs of co-stimulatory molecule genes including: four SNPs of CTLA4 gene (rs5742909, 162 bp upstream from the first nucleotide of the 5’-UTR, also known as -318 C.T, rs733618, -1722 T/C, rs4553808, -1661 A.G, an exonic SNP, rs231775, exon 1, Thr17Ala, also known as +49A/G), two SNPs from the PDCD1 gene (PDCD.1.3 rs11568821 A/G intron SNP, PDCD.1.9 rs2227982C.T exon SNP), one SNP from the ICOS gene (1720 C/T) and one SNP from the CD28 (rs3116496, IVS3 +17 C/T) gene in chromosome 2q33-37. Polymerase chain reaction (PCR) cycles, the PCR thermocycling condition, primers and types of PCR for the co-stimulatory molecules are summarized in Table 1. Genetic polymorphisms for PDCD.1.3 A/G, PDCD.1.9 C/T, CD28 +17 C/T, CTLA4 -1722 T/C, -1661 A/G, -318 C/T, + 49 A/G and ICOS +1720 C/T were analyzed using the PCR-RFLP method, as previously described (34-39).
| SNP | Primer | Thermo Cycling Condition | Product Size | Method |
|---|---|---|---|---|
| CTLA4, -1661A/G | 1 cycle: 95°C 5 min; 35 cycles: 95°C 50 sec, 59°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 7 min | A allele: 139,347 bp; G allele:486 bp | MseI based RFLP | |
| F | 5-CTAAGAGCATCCGCTTGCACCT-3 | |||
| R | 5-TTGGTGTGATGCACAGAAGCCTTTT-3 | |||
| CTLA4, -318 T/C | 1 cycle: 94°C 5 min; 35 cycles: 94°C 30 sec, 58°C 30 sec, 72°C 30 sec; 1 cycle: 72°C 7 min | C allele: 21 + 226 bp; T allele: 96+130 bp | MseI based RFLP | |
| F | 5-AAATGAATTGGACTGGATGG-3 | |||
| R | 5-TTACGAGAAAGGAAGCCGTG-3 | |||
| CTLA4, +49A/G | 1 cycle: 95°C 4 min; 30 cycles: 94°C 60 sec, 57°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | A allele: 162 bp; G allele: 88,78 bp | BbvI based RFLP | |
| F | 5-GCTCTACTTCCTGAAGACCT-3 | |||
| R | 5-AGTCTCACTCACCTTTGCAG-3 | |||
| CTLA4, -1722T/C | 1 cycle: 95°C 5 min; 30 cycles: 95°C 60 sec, 62°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | T allele: 486 bp; C allele: 216,270 bp | BbvI based RFLP | |
| F | 5-CTAAGAGCATCCGCTTGCACCT-3 | |||
| R | 5-TTGGTGTGATGCACAGAAGCCTTTT-3 | |||
| CD28, +17 C/T | 1 cycle: 95°C 5 min; 30 cycles: 95°C 60 sec, 58°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | C allele: 147 bp; T allele: 22,125 bp | Eco47III based RFLP | |
| F | 5-TTTTCTGGGTAAGAGAAGCAGCGC-3 | |||
| R | 5-GAACCTACTCAAGCATGGGG-3 | |||
| PD.1.9, -7625T/C | 1 cycle: 94°C 5 min; 30 cycles: 94°C 60 sec, 58°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | T allele: 174,234 bp; C allele: 174,89,145 bp | Bpu10I based RFLP | |
| F | 5-GGACAGCTCAGGGTAAGCAG-3 | |||
| R | 5-AGGGTCTGCAGAACACTGGT-3 | |||
| PD.1.3, -7146A/G | 1 cycle: 95°C 4 min; 30 cycles: 95°C 60 sec, 55.5°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | G allele: 180 bp; A allele: 57,123 bp | PstI based RFLP | |
| F | 5-CCCCAGGCAGCAACCTCA-3 | |||
| R | 5-GACCGCAGGCAGGCACAT-3 | |||
| ICOS, 1720 C/T | 1 cycle: 95°C 5 min; 30 cycles: 95°C 60 sec, 58°C 60 sec, 72°C 60 sec; 1 cycle: 72°C 5 min | ´ C allele: 824 bp; T allele: 304,520 bp | NcoI based RFLP | |
| F | 5-TTACCAAGACTTTAGATGCTTTCTT-3 | |||
| R | 5-GAATCTTTCTAGCCAAATCATATTC-3 |
The Thermo Cycling Condition, Primers and Types of PCR for the ICOS, CD28, PD.1 and CTLA4 Co-stimulatory Molecules
3.3. Cytomegalovirus Antigenemia Assay
The CMV active infection was evaluated in EDTA-treated whole blood samples collected from kidney transplant patients by the CMV antigenemia method using CMV Brite Turbo kit (IQ Products, Groningen, The Netherlands), according to manufactures’ instruction.
3.4. Hepatitis C Virus Enzyme Linked Immunosorbent Assay
The history of HCV infection was evaluated using the HCV antibody assay on plasma samples of kidney transplant patients by a third generation ELISA kit (DIAPRO, Italy), according to manufacturer’s instruction.
3.5. Statistical Analysis
Allele and genotype frequencies were calculated in studied kidney transplant patients by direct gene counting. Analysis was carried out using the statistical package for the social sciences (SPSS), version 16. The frequencies of the alleles and genotypes were compared in the two groups by chi-square test and Fisher’s exact test. Odds ratios and 95% confidence intervals (CIs) for relative risks were calculated. A probability value of P < 0.05 was considered as statistically significant and all the reported P values were two-tailed. The LD2SNPing program V 2.0 (http//.www.bio.kuas.edu.tw.LD2SNPing) was used to estimate linkage disequilibrium (LD) and the results were evaluated using Arlequin V311.
4. Results
4.1. Patient’s Profile
The age range of kidney transplant patients was between 10 and 79 years old with mean of 38.28 ± 14.29. Overall, 113 of 172 (65.70%) kidney transplant recipients were male and 59 of 172 (34.30%) were female with a mean age of 38.28 ± 14.29 years old. Forty-five (26.16%) transplant patients were shown acute rejection and 127 (73.84%) patients did not experience acute rejection. Acute rejection was confirmed in 26 of 45 (57.8%) male and 19 of 45 (42.2%) female patients. Ninety-nine of 172 (57.6%) kidney transplant patients received transplant from cadaver and 73 (42.4 %) received transplant from living donors. Thirty-two of 99 (32.3%) who had received a transplant from cadaver and 13 of 73 (17.8%) with living donors experienced acute rejection.
4.2. Viruses and Kidney Transplantation
CMV active infection was found in 31 of 172 (18.02%) patients with an age range between 18 and 76 years with mean 36.90 ± 15.35. The 18 of 31 (58.06%) CMV infected patients were male and the rest (13 of 31; 41.94%) were female. Furthermore, 23 of 31 (74.19%) CMV infected patients received a graft from cadaver and 8 of 31 (25.81%) received a kidney from a living donor. Twenty of 31 (64.55%) CMV-infected patients experienced acute rejection and 11 of 31 (35.45%) CMV infected patients did not experience acute rejection. HCV infection was only found in two of 172 (1.16%) kidney transplant patients, who were male and received a graft from cadaver. One of the HCV-infected patients experienced acute rejection. No significant relationship was found between polymorphisms of co-stimulatory molecules and HCV infection in any kidney transplant patient.
4.3. Co-stimulatory Molecule Gene Polymorphisms and Cytomegalovirus Infection
Genotype frequencies of CD28, ICOS, CTLA4 and PDCD1 costimulatory genes in acute rejected and non-acute rejected kidney transplant patients with CMV infection are presented in Table 2. Significant associations were found between CTLA4 1722 TT and TC gene polymorphisms and also T allele with acute rejection in CMV-infected kidney transplant patients (P = 0.04, OR = undefined, 95% CI = undefined; P = 0.04, OR = 0.001, 95% CI = 0.001 - 2.24; P = 0.05, OR = undefined, 95% CI = undefined, respectively). A significant association was also found between the T allele of CD28 +17 C/T polymorphism with acute rejection in CMV-infected kidney transplant patients (P = 0.05, OR = 3.08, 95% CI = 0.84 - 11.84) and also significant associations were found between CD28 +17 C/T TT genetic polymorphism and non-acute rejection in CMV infected kidney transplant patients (P = 0.03, OR = 0.19, 95% CI = 0.03 - 1.18), (Table 2). On the other hand, genotype frequencies of CD28, ICOS1720, CTLA4 and PD.1 co-stimulatory molecules between male and female kidney transplant patients with CMV infection are presented in Table 2. A significantly higher frequency of CTLA4 49A/G AA genotype and A allele polymorphism were found in CMV infected female patients (P = 0.02, OR = 0.15, 95% CI = 0.02 - 1.05; P = 0.01, OR = 0.17, 95% CI = 0.02 - 0.93, respectively) and also a significantly higher frequency of PDCD-1.3 GG genotype and G allele polymorphisms was found in CMV infected female patients (P = 0.02, OR = 0.00, 95% CI = 0.00 - 1.11; P = 0.005, OR = 0.00, 95% CI = 0.00 - 0.69, respectively) (Table 3).
| SNP | Rejection a | Non-Rejection a | P Value b | OR | 95% Cl |
|---|---|---|---|---|---|
| CTLA-4, -318 C/T | |||||
| Genotypes | |||||
| CC | 15 (75) | 8 (66.7) | 0.88 | 1.13 | 0.16 - 7.83 |
| CT | 5 (25) | 3 (27.3) | 0.88 | 0.89 | 0.13 - 6.39 |
| TT | 0 (0) | 0 (0) | Undefined | Undefined | Undefined |
| Alleles | 0.89 | 1.11 | 0.18 - 6.21 | ||
| C | 35 (87.5) | 19 (87.5) | |||
| T | 5 (12.5) | 3 (13.6) | |||
| CTLA-4, +49 A/G | |||||
| Genotypes | |||||
| AG | 6 (30) | 4 (36.4) | 0.71 | 0.75 | 0.12 - 4.63 |
| AA | 13 (65) | 6 (17.6) | 0.56 | 1.55 | 0.27 - 9.03 |
| GG | 1 (5) | 1 (54.5) | 0.65 | 0.53 | 0.01 - 21.97 |
| Alleles | 0.51 | 1.50 | 0.38 - 5.93 | ||
| A | 32 (87.5) | 16 (72.7) | |||
| G | 8 (12.5) | 6 (27.3) | |||
| CTLA-4, -1661 A/G | |||||
| Genotypes | |||||
| GG | 1 (5) | 0 (0) | 0.45 | Undefined | Undefined |
| AA | 14 (70) | 7 (63.6) | 0.71 | 1.33 | 0.22 - 8.25 |
| GA | 5 (25) | 4 (36.4) | 0.50 | 0.58 | 0.09 - 3.73 |
| Alleles | 0.94 | 1.05 | 0.22 - 4.80 | ||
| A | 33 (82.5) | 18 (81.8) | |||
| G | 7 (17.5) | 4 (18.2) | |||
| CTLA-4, -1722 C/T | |||||
| Genotypes | |||||
| TT | 20 (100) | 9 (81.8) | 0.04 c | Undefined | Undefined |
| TC | 0 (0) | 2 (18.8) | 0.04 c | 0.001 | 0.001 - 2.24 |
| CC | 0 (0) | 0 (0) | Undefined | Undefined | Undefined |
| Alleles | 0.05 c | Undefined | Undefined | ||
| T | 40 (100) | 20 (90.9) | |||
| C | 0(0) | 2 (9.1) | |||
| PDCD-1, 1.9T/C | |||||
| Genotypes | |||||
| CC | 14 (70) | 10 (90.9) | 0.18 | 0.23 | 0.01 - 2.63 |
| TC | 5 (25) | 1 (9.9) | 0.28 | 3.33 | 0.28 - 87.48 |
| TT | 1 (5) | 0 (0) | 0.45 | Undefined | Undefined |
| Alleles | 0.14 | 4.45 | 0.48 - 103.32 | ||
| T | 7 (17.5) | 1 (4.5) | |||
| C | 33 (82.5) | 21 (95.5) | |||
| PDCD-1, 1.3A/G | |||||
| Genotypes | |||||
| GG | 16 (80) | 9 (81.8) | 0.90 | 0.89 | 0.09 - 7.70 |
| GA | 2 (10) | 1 (9.1) | 0.93 | 1.11 | 0.06 - 35.48 |
| AA | 2 (10) | 1 (9.1) | 0.93 | 1.11 | 0.06 - 35.48 |
| Alleles | 0.88 | 0.89 | 0.15 - 4.75 | ||
| G | 34 (85) | 19 (84.4) | |||
| A | 6 (15) | 3 (13.6) | |||
| CD28 +17 C/T | |||||
| Genotypes | |||||
| CC | 4 (20) | 1 (9.1) | 0.42 | 2.50 | 0.20 - 67.85 |
| TC | 11 (55) | 3 (27.3) | 0.13 | 3.26 | 0.53 - 22.00 |
| TT | 5 (25) | 7 (63.6) | 0.03 c | 0.19 | 0.03 - 1.18 |
| Alleles | 0.05 c | 3.08 | 0.84 - 11.84 | ||
| C | 19 (47.5) | 5 (22.7) | |||
| T | 21 (52.5) | 17 (77.3) | |||
| ICOS 1720 C/T | |||||
| Genotypes | |||||
| CT | 6 (30) | 4 (36.4) | 0.71 | 0.75 | 0.12 - 4.63 |
| CC | 13 (65) | 7 (63.6) | 0.93 | 1.06 | 0.18 - 6.33 |
| TT | 1 (5) | 0(0) | 0.45 | Undefined | Undefined |
| Alleles | 0.86 | 0.89 | 0.19 - 3.94 | ||
| C | 32 (80) | 18 (81.8) | |||
| T | 8 (20) | 4 (18.2) | |||
| SNP | CMV Infected | CMV Infected | CMV Infected | CMV Non-Infected | CMV Non-Infected | CMV Non-Infected | P Value 1 b,c | P Value 2 b,d | P Value 3 b,e |
|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Total | ||||
| CTLA4, 1661 A/G | |||||||||
| Genotypes | |||||||||
| GG | 1 (5.5) | 0 (0) | 1 (3.2) | 5 (5.3) | 5 (10.9) | 10 (7.1) | 0.42 | 0.95 | 0.21 |
| AA | 11 (61.1) | 11 (84.6) | 22 (71) | 57 (60) | 25 (54.3) | 82 (58.1) | 0.18 | 0.92 | 0.04 f |
| GA | 6 (33.3) | 2 (15.4) | 8 (25.8) | 33 (34.7) | 16 (34.8) | 49 (34.8) | 0.33 | 0.91 | 0.17 |
| Alleles | |||||||||
| G | 8 (22.2) | 2 (7.7) | 10 (16.1) | 43 (22.6) | 26 (28.3) | 69 (24.5) | 0.15 | 0.95 | 0.02 f |
| A | 28 (77.8) | 24 (92.3) | 52 (83.9) | 147 (77.4) | 66 (71.7) | 213 (75.5) | |||
| CTLA4, 1722 T/C | |||||||||
| Genotypes | |||||||||
| TT | 16 (88.9 ) | 13 (100) | 29 (93.5) | 85 (89.5) | 42 (91.4) | 127 (90.1) | 0.40 | 0.94 | 0.27 |
| TC | 2 (11.1) | 0 (0) | 2 (6.5) | 9 (9.5) | 2 (4.3) | 11 (7.8) | 0.79 | 0.82 | 0.44 |
| CC | 0 (0) | 0 (0) | 0 (0) | 1 (1) | 2 (4.3) | 3 (3) | 0.41 | 0.66 | 0.44 |
| Alleles | |||||||||
| T | 34 (94.4) | 26 (100) | 60 (96.8) | 179 (94.2) | 86 (93.5) | 265 (94) | 0.38 | 0.95 | 0.18 |
| C | 2 (5.6) | 0 (0) | 2 (3.2) | 11 (5.8) | 6 (6.5) | 17 (6) | |||
| CTLA4, -318 C/T | |||||||||
| Genotypes | |||||||||
| CC | 13 (72.2) | 10 (76.9) | 23 (74.2) | 76 (80) | 36 (78.3) | 112 (79.5) | 0.52 | 0.45 | 0.91 |
| TC | 5 (27.8) | 3 (23.1) | 8 (25.8) | 16 (16.8) | 8 (17.4) | 24 (17) | 0.25 | 0.27 | 0.64 |
| TT | 0 (0) | 0 (0) | 0 (0) | 3 (3.2) | 2 (4.3) | 5 (3.5) | 0.28 | 0.44 | 0.44 |
| Alleles | |||||||||
| C | 31 (86.1) | 23 (88.5) | 54 (87.1) | 168 (88.4) | 80 (87) | 248 (87.9) | 0.53 | 0.69 | 0.83 |
| T | 5 (13.9) | 3 (11.5) | 8 (12.9) | 22 (11.6) | 12 (13) | 34 (12.1) | |||
| CTLA4, +49 A/G | |||||||||
| Genotypes | |||||||||
| AG | 8 (44.4) | 2 (15.4) | 10(32.2) | 30 (31.6) | 17 (37) | 47 (33.4) | 0.90 | 0.28 | 0.14 |
| AA | 8 (44.4) | 11 (84.6) | 19 (61.3) | 51 (53.7) | 28 (60.8) | 79 (56) | 0.59 | 0.47 | 0.11 |
| GG | 2 (11.1) | 0 (0) | 2 (6.5) | 14 (14.7) | 1 (2.2) | 15 (10.6) | 0.47 | 0.68 | 0.59 |
| Alleles | |||||||||
| A | 24 (66.7) | 24 (92.3) | 48 (77.4) | 132 (69.5) | 73 (79.3) | 205 (72.7) | 0.44 | 0.73 | 0.12 |
| G | 12 (33.3) | 2 (7.7) | 14 (22.6) | 58 (30.5) | 19 (20.7) | 77 (27.3) | |||
| PDCD-, 1.3 A/G | |||||||||
| Genotypes | |||||||||
| GG | 12 (66.6) | 13 (100) | 25 (80.6) | 65 (68.4) | 34 (73.9) | 99 (70.2) | 0.24 | 0.88 | 0.03 f |
| GA | 3 (16.7) | 0 (0) | 3 (9.7) | 24 (25.3) | 8 (17.4) | 32 (22.7) | 0.10 | 0.43 | 0.10 |
| AA | 3 (16.7) | 0 (0) | 3 (9.7) | 6 (6.3) | 4 (8.7) | 10 (7.1) | 0.62 | 0.13 | 0.20 |
| Alleles | |||||||||
| G | 27 (75) | 26 (100) | 53 (85.5) | 154 (81.1) | 76 (82.6) | 230 (82.1) | 0.46 | 0.40 | 0.02 f |
| A | 9 (25) | 0 (0) | 9 (14.5) | 36 (18.9) | 16 (17.4) | 52 (18.4) | |||
| PDCD-, 1.9 C/T | |||||||||
| Genotypes | |||||||||
| CC | 15 (83.3) | 9 (69.2) | 24 (77.4) | 71 (74.7) | 33 (71.7) | 104 (73.7) | 0.67 | 0.43 | 0.86 |
| CT | 3 (16.7) | 3 (23.1) | 6 (19.3) | 13 (13.7) | 8 (17.4) | 21 (14.9) | 0.53 | 0.73 | 0.64 |
| TT | 0 (0) | 1 (7.7) | 1 (3.2) | 11 (11.6) | 5 (10.9) | 16 (11.3) | 0.17 | 0.12 | 0.73 |
| Alleles | 0.27 | 0.13 | 0.96 | ||||||
| C | 33 (91.7) | 21 (80.8) | 54 (87.1) | 155 (81.6) | 74 (80.4) | 229 (81.2) | |||
| T | 3 (8.3) | 5 (19.2) | 8 (12.9) | 35 (18.4) | 18 (19.6) | 53 (18.8) | |||
| CD28 +17 C/T | |||||||||
| Genotypes | |||||||||
| CC | 4 (22.2) | 1 (7.7) | 5 (16.1) | 17 (17.9) | 9 (19.6) | 26 (18.4) | 0.76 | 0.66 | 0.31 |
| TC | 7 (38.9) | 7 (53.8) | 14 (45.2) | 32 (33.7) | 14 (30.4) | 46 (32.6) | 0.18 | 0.67 | 0.11 |
| TT | 7 (38.9) | 5 (38.5) | 12 (38.7) | 46 (48.4) | 23 (50) | 69 (50) | 0.30 | 0.45 | 0.46 |
| Alleles | 0.55 | 0.42 | 0.98 | ||||||
| C | 15 (41.7) | 9 (34.6) | 24 (38.7) | 66 (34.7) | 32 (34.8) | 98 (34.7) | |||
| T | 21 (58.3) | 17 (65.4) | 38 (61.3) | 124 (65.3) | 60 (62.2) | 184 (62.3) | |||
| ICOS, 1720 C/T | |||||||||
| Genotypes | |||||||||
| CT | 8 (44.4) | 2 (15.4) | 10 (32.2) | 38 (40) | 11 (23.9) | 49 (34.7) | 0.79 | 0.72 | 0.51 |
| CC | 10 (55.6) | 10 (76.9) | 20 (64.6) | 53 (55.8) | 31 (67.4) | 84 (59.6) | 0.61 | 0.98 | 0.50 |
| TT | 0(0) | 1 (7.7) | 1 (3.2) | (4.2) | 4 (8.7) | 8 (5.7) | 0.57 | 0.37 | 0.90 |
| Alleles | 0.52 | 0.79 | 0.54 | ||||||
| C | 28 (77.8) | 22 (84.6) | 50 (80.6) | 144 (75.8) | 73 (79.3) | 217 (77) | |||
| T | 8 (22.2) | 4 (15.4) | 12 (19.4) | 46 (24.2) | 19 (20.7) | 65 (23) | |||
The Genotypes and Allele Frequencies of CTLA4, PD.1, CD28 and ICOS Co-stimulatory Molecules Between CMV Infected and Non-Infected Kidney Transplant Patientsa
Moreover, the frequency of CTLA4, PD.1, CD28 and ICOS genotypes and alleles in CMV infected and non-infected kidney transplant patients are presented in Table 3. The AA genotype and A allele of CTLA4 1661 A/G had a higher frequency in CMV non-infected female patients compared with CMV infected ones (P = 0.04 and P = 0.02 respectively). The G allele and GG genotype of PDCD-1.3 had higher frequency in CMV non-infected female patients (P = 0.03 and P = 0.02 respectively), (Table 3).
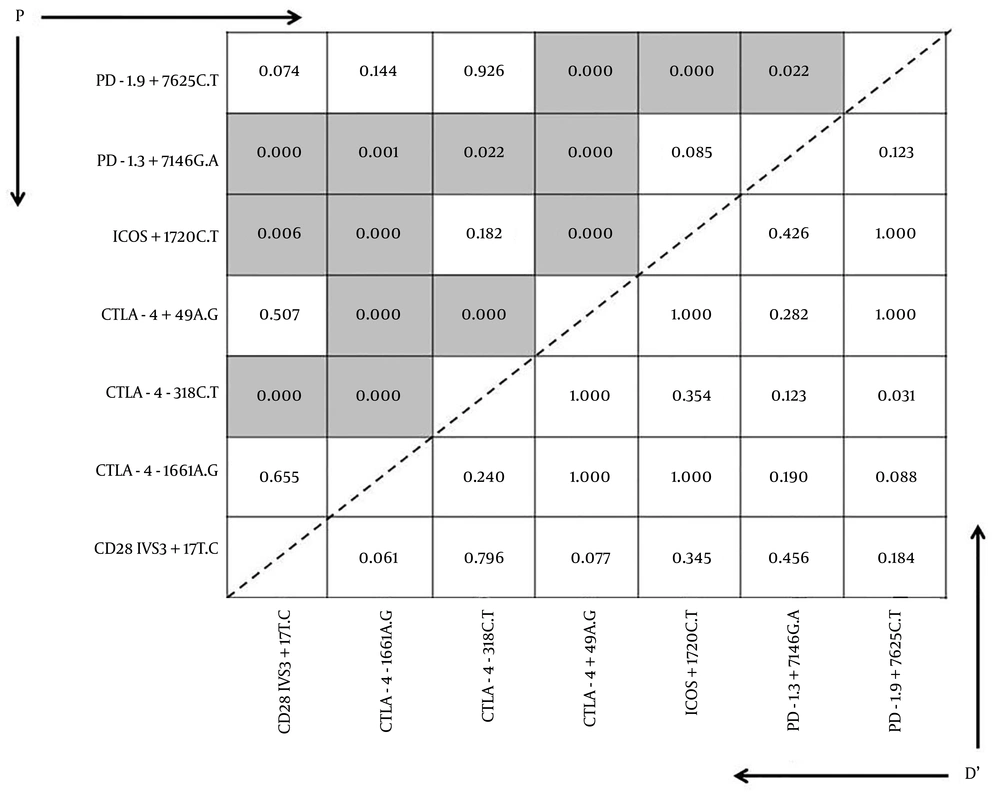
4.4. Linkage Disequilibrium Between Cytomegalovirus Infection and Costimulatory Molecules
A linkage disequilibrium was found in CMV infected patients between CTLA4 (+49) and PD-1 + 7625 and ICOS + 1720 CTLA4 (-1661) gene polymorphisms (D’ = 1.000, P < 0.000), and between ICOS + 1720 and CTLA4 (-1661) and PD-1 + 7625 gene polymorphisms (D’ = 1.000, P < 0.000), and also found between CTLA4 (-318) and CD28 gene polymorphism (D’= 0.796, P < 0.000), (Figure 1).
4.5. Co-stimulatory Molecule Gene Polymorphisms and Risk Factors
The mean ages of the patients, who had experienced an acute rejection episode after three months of follow-up was 36.16 ± 12.97. A significant association was found in A+ blood group with higher frequency of CTLA4 1661 A/G and -318 C/T gene polymorphisms in transplant patients (P = 0.05 and P = 0.02, respectively). Also, a significant association was found between age and acute rejection in kidney transplant patients (P = 0.04).
5. Discussion
Patients with CMV appear to have reduced allograft survival (40). Whether this is a direct effect on the allograft or an indirect consequence of infection leading to acute tubular necrosis and cellular rejection has been disputed. A few studies have shown the presence of CMV inclusions in allograft tissue, confounding the association of CMV and transplant glomerulonephropathy (4, 41). The negative role of CMV infection in inducing acute and chronic kidney transplant outcomes remains controversial, yet several studies suggested an immune response-based association between CMV pathogenesis and allograft rejection (36, 42, 43). Regarding immune system tools, costimulatory molecules and their related gene polymorphisms may affect post-transplant virus-related outcomes (22, 23). Therefore, in this study the role of co-stimulatory molecules’ genetic polymorphisms that may have influenced viral pathogenesis were evaluated in kidney transplant patients. The CD28 and ICOS are major co-stimulatory molecules activate T cell related pathways and introduce immune responses yet CTLA4 and PDCD1 negatively regulate T cell reactivation against viral infections (19, 20). Several genetic polymorphisms that occurred in co-stimulatory molecules encourage or reduce inflammatory T cell-based immune responses in transplant patients (21).
The PDCD-1 was expressed increasingly in the viral infected population, especially in patients with symptomatic CMV disease (44). However, in healthy asymptomatic CMV-infected individuals, levels of PDCD-1 on CMV-specific CD8 T cells were low similar to HIV-infected individuals (26). The simultaneous role of anti-rejection therapy in up-regulation of PDCD-1 was ruled out in CMV infected patients (26). In our earlier reports significant associations were not found between PDCD-1 genetic polymorphisms with active CMV infections in the liver and also hematopoietic stem cell transplant patients (24, 45). Similarly, in this study also the G allele and GG genotype of PDCD-1.3 had significantly higher frequency in CMV non-infected female patients. The CTLA4 has an important role in immunologic homeostasis, as a negative regulator of T cell activation (46).
Earlier reports present different controversial associations between CTLA4 genetic polymorphisms and viral infections and transplant outcomes. In one study, higher frequencies of CTLA4 318 C/T CT genotype and C allele and AG genotype of CTLA4 +49 A/G was found in CMV infected acute rejected liver transplant patients (24). Patients receiving hematopoietic stem cell transplant (HSCT) from a donor with significant higher presentation of CTLA4 CT60 AA genotype compared with AG/GG genotypes experienced lower grades of acute Graft versus host disease (aGVHD), yet experienced early post-transplant CMV infection (47). In our earlier study, a significant association was also found between CTLA4 +49AG AA genotype and A allele, found in CMV infected female liver transplant patients. The outcome of HBV infection was reduced in patients with higher frequency of GG and CC genotypes of CTLA4 +49 A/G and 1722 T/C loci, respectively (29, 48).
Successful clearance of HCV infection during therapy appeared by reduction of CTLA4 levels, which was encouraged with higher presentation of GG and CC genotypes of CTLA4 +49 A/G and -318 C/T loci, respectively (28). These findings present the determinative role of CTLA4 polymorphisms on control of T cell proliferation leading to recovery of patients from chronic viral hepatitis clinical complications. Similarly, in this report, a significantly higher frequency of CTLA4 1722 TT and TC genotypes and also T allele were found in acute rejected CMV infected kidney transplant patients. Also significantly higher presentation of CTLA4 49AG AA genotype and A allele were found in CMV infected female patients.
The CD28 was the first cell surface co-stimulatory protein, which was diagnosed with the ability to activate T cell proliferation and development (49, 50). The CD28 T cell activation pathway plays a key role in immune responses against viral infections and in graft rejection (50). In our earlier report significantly higher frequency of the C allele and CC genotype of the CD28+17C/T gene locus was found in hepatitis B virus (HBV) infected allogeneic HSCT patients (24). Similarly, in our other study significantly higher frequency of T allele and TT genotype of the CD28 +17 C/T locus was found in CMV infected allogeneic HSCT patients with aGVHD (45). On the other hand, Kusztal et al. did not found any association between CD28+17C/T genetic polymorphisms and acute rejection in kidney transplant patients (11). In another study, no association was found between acute kidney graft rejections with CD28+17C/T genetic polymorphisms (51).
In this study, significantly higher frequency of the T allele of CD28 +17 C/T genetic polymorphism was observed in acute rejected and CMV infected kidney transplant patients. The ICOS gene, which is one of the CD28 family is expressed in resting T cells and activates T cells and CD28 co-stimulatory molecules, enhanced by cytokines IL12 and IL23 (52). In an earlier study an association between ICOS gene polymorphism and rejection was presented in kidney transplant patients (42). However, in this study no significant associations were found between ICOS1720 C/T genetic polymorphisms and CMV infection in kidney transplant patients. In conclusion, based on these findings, CTLA4 and CD28 genetic polymorphisms regulate T cell activation and influence active CMV infection in kidney transplant patients. This study had a small group of evaluated transplant patients, thus it is better to confirm these results by further investigations.