1. Background
The hepatitis C virus (HCV) is a single-stranded RNA virus belonging to the family Flaviviridae and genus hepacivirus (1). Currently, an estimated 130–150 million people worldwide are infected with HCV, about 2 - 3% of the world population (2). Interferon (IFN)-alfa and ribavirin (pegylated) is currently the standard of care treatment (3), but new direct-acting antiviral treatments (boceprevir and telaprevir) have recently received FDA approval (4). Treatment response rates differ significantly among infected patients according to viral genotype. Up to 80% of genotype 2- and 3-infected patients and 40 - 50% of patients infected with genotype 1 can be cured (3).
Recently, it has been reported that host factors and several genetic factors are also correlated to IFN therapeutic effects (5-7). One of the most important genetic factors is polymorphisms in the interleukin (IL) 28-B gene region. IL28B (IFN-λ3) encodes interferon-λ3 and is located on the long arm of chromosome 19, on 19q13.13; it contains six exons (8). IFN-λ3 has been reported to be involved in the suppression of HCV replication (9). Based on genome-wide associated studies, several single nucleotide polymorphisms (SNPs) located in and around the IL28B gene were found to be associated with treatment response in patients chronically infected with HCV (10). One of the most important SNPs, the rs12979860 polymorphism located in 3181 bp upstream of the IL28B gene and specified by the C or T allele, is strongly associated with sustained virological response (SVR). This polymorphism may explain much of the difference in response between different population groups.
2. Objectives
The main aim of this study was to determine the allelic and genotypic frequencies of the IL28B rs12979860 polymorphism in SVR patients in the Iranian population compared with patients who did not respond to treatment. The study also investigated whether there was a significant association between the viral load and the IL28B rs12979860 polymorphism.
3. Methods
3.1. Patients
This cross-sectional study was carried out on samples from patients suspected of HCV infection. Samples were referred to the Bou-Ali pathobiology laboratory, as the reference molecular laboratory in Yazd, Iran, from March 2010 to June 2013. The research protocol was approved by the ethics committee of Yazd Shahid Sadoughi University of Medical Sciences. Signed informed consent was obtained from each patient, and all samples were examined by the gastroenterologist. The patients were of Caucasian ancestry and did not have apparent autoimmune hepatitis, alcoholic liver disease, primary biliary cirrhosis, sclerosing cholangitis, Wilson’s disease, α1-antitrypsin deficiency, decompensated cirrhosis or overt hepatic failure or a current or past history of alcohol abuse, previous liver transplantation or evidence of hepatocellular carcinoma.
Five milliliters of peripheral blood samples were taken and centrifuged after clotting, and the serum was harvested and stored at -70°C until used. The patients’ serum was screened for the hepatitis B surface antigen (HBs Ag) using a chemiluminescence assay (LIAISON DiaSorin SpA, Italy). Human immunodeficiency virus (HIV) and HCV antibodies were determined using a commercial enzyme-linked immunosorbent assay kit (Diapro, Diaplus, Italy). Positive samples for HBsAg and anti-HIV were excluded from this study. Seropositive subjects for HCV antibodies were analyzed using the real-time polymerase chain reaction (RT-PCR) method.
The blood viral load was determined in accordance with the manufacturer’s instructions (AJ Roodscreen, GmbH). The Rotor-Gene 6000 (Corbett Research, Sydney, Australia) was used to determine the viral load level. The analytical sensitivity of the kit was 200 copies/ml. HCV-RNA-positive samples were genotyped using the HCV genotypes kit (Sacace Biotechnologies S.r.l, Italy). The kit was able to determine HCV genotypes 1a, 1b, 2, 3 and 4. All patients with detected virus in their serum were treated with peginterferon alfa-2a (180 µg/week) subcutaneously plus weight-based ribavirin (1000 mg d-1 for weight < 80 kg and 1200 mg d-1 for weight > 80 kg). Patients with genotype 1 HCV were treated for 48 weeks and patients with genotype 3 for 24 weeks.
Six months after the end of treatment, the subjects were referred to the Bou-Ali pathobiology laboratory. Five milliliters of peripheral blood samples were taken from all patients; 2 mL of each sample was collected in an EDTA-treated tube for DNA extraction, and the reminder was centrifuged after clotting and the serum harvested and stored at -70°C until used. The viral load in the serum was evaluated. The samples were divided, according to the presence or absence of HCV in the blood, into a responder group (group 1) with no detectable viral load and a nonresponder group (group 2) with a detectable load of virus in the serum. Group 1 consisted of 45 patients, and group 2 consisted of 30 patients.
3.2. Genetic Analysis
Genotyping for the IL28B rs12979860 polymorphism was performed using the polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method. Genomic DNA was extracted from whole blood samples using a commercial kit (AccuPrep Genomic DNA Extraction Bioneer Kit, Republic of Korea). Primers were designed according to published sequences in the Genome Database (GenBank), using the software PRIMER3.
The information for the primer sequences is as follows:
Forward 5’-GCTTATCGCATACGGCTAGG-3’ and
Reverse 5’-AGGCTCAGGGTCAATCACAG-3’.
PCR amplification was performed in a total volume of 25 μL containing 4 μL of DNA, 12.5 μL master mix (Ampliqon, Denmark), 6 μL distilled water and 1.25 μL each of specific reverse and forward primers (Macro-gene). Samples containing genomic DNA were subjected to an initial denaturation at 94°C for 5 minutes followed by 40 cycles of denaturation at 94°C for 45 seconds, annealing at 62°C for 45 seconds and elongation at 72°C for 30 seconds. A final extension was carried out for 5 minutes at 72°C (Applied Biosystems, ABI, Foster City, CA, USA). PCR products were subjected to electrophoresis in a 2% agarose gel stained with green viewer. Then the PCR product was digested by the BstU-I restriction enzyme according to kit instructions (New England Biolabs, Hitchin, UK). Fragments were analyzed by electrophoresis in a 3% agarose gel staining with green viewer. All the gels were imaged using an E-gel imager (Life Technologies) instrument. Five samples in each group were randomly selected to perform the repeated assay, and the results were 100% concordant.
3.3. Statistical Analysis
Statistical analysis was performed using SPSS version 16 (SPSS Inc., Chicago, IL, USA). SNPs were evaluated by chi-square test for deviation from Hardy-Weinberg equilibrium. The genotype and allele frequencies of SNPs rs12979860 were calculated by direct count. The differences in allele and genotype frequencies between the two groups were determined using a chi-square test. We used an independent sample t-test to determine the association between the viral load of with IL28B rs12979860 polymorphism and to determine the association between the mean viral load and the virus genotypes. P < 0.05 were considered statistically significant.
4. Results
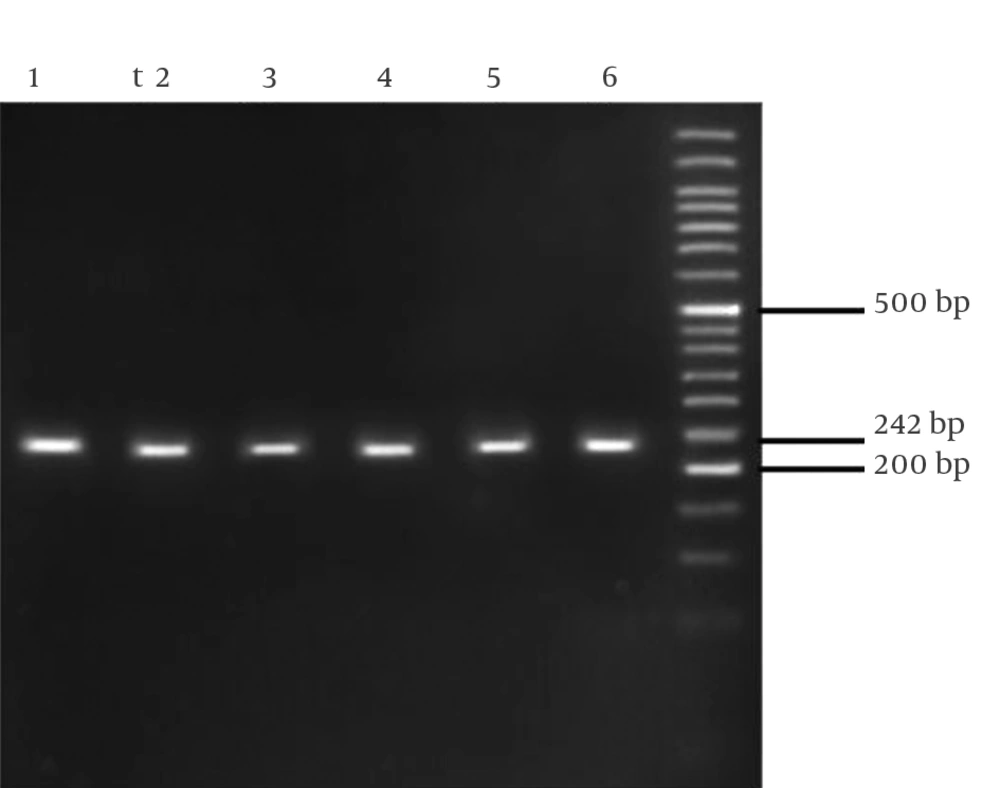
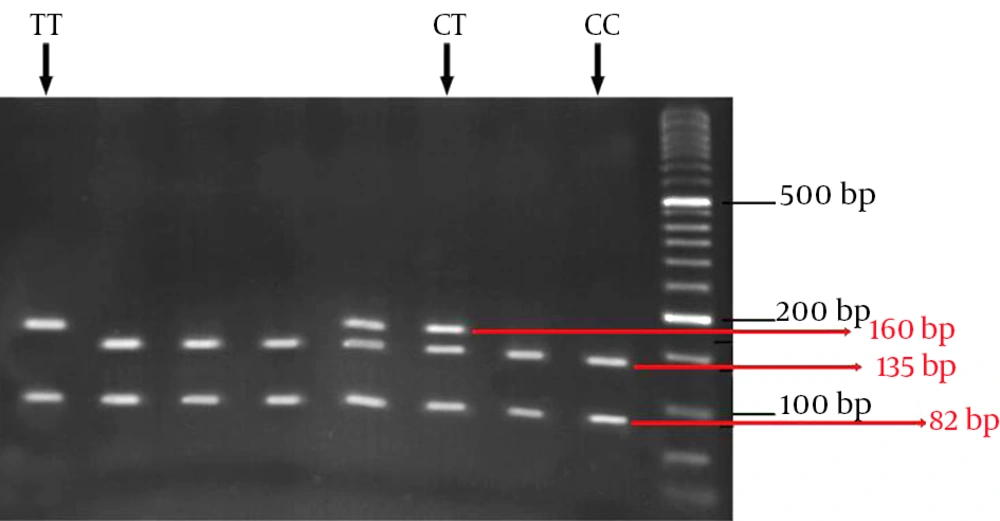
In this study, 75 HCV infected patients, including 45 responders to treatment (group 1) and 30 nonresponders (group 2), were genotyped for the IL28B rs12979860 polymorphism. The IL28B genotype frequencies were in Hardy-Weinberg equilibrium in the two groups. Characteristics of the studied patients are shown in Table 1. There was no significant difference in age between the two groups (P = 0.5). The undigested PCR product size was 242 bp for SNP rs12979860 (Figure 1). Restriction digestion for the CC genotype generated 135, 82 and 25 bp fragments, whereas the CT genotype generated 160, 135, 82 and 25 bp and the TT genotype produced 160 and 82 bp fragments (Figure 2).
The genotype frequencies of rs12979860 polymorphism in the responder group were CC (28.9%), CT (37.8%) and TT (33.3%) and in the non-responder group were CC (6.7%), CT (43.3%) and TT (50%). Genotype distributions for the IL28B polymorphism were significantly different between responder and nonresponder patients (P = 0.03). Patients with the homozygous CC genotype had a significantly higher rate of response to antiviral therapy than those with the TT or CT genotypes. The frequency of allele T was 52.2% in the responder group and 71.7% in the nonresponder group. The T allele frequency was significantly higher in the nonresponder group compared with the responder group (P = 0.02). Allele and genotype frequencies in the two groups are presented in Table 2.
aP = 0.55.
bP = 0.9.
aValues are expressed as No. (%).
bP = 0.02.
cP = 0.03.
The mean viral load in all patients was 855941 ± 178482 copies/mL. In group 1, the mean viral load was 845433 ± 227081 copies/mL, and it was 871700 ± 292990 copies/mL in group 2. The mean viral load between the two groups was not significant (P = 0.9). There was no association between the viral load and IL28B rs12979860 in either the responder (P = 0.3) or the nonresponder groups (P = 0.2).
The genotypes in all samples were either 1 or 3. In 25 (55.6%) patients of group 1 and 22 (73.3%) patients of group 2, genotype 1 was determined, while the rest showed genotype 3. There was no significant association between the genotype of the virus and rs12979860 IL28B polymorphism (Table 3). Nor were there any significant differences between the mean viral load and the virus genotypes in either group 1 (P = 0.6) or group 2 (P = 0.8).
5. Discussion
In patients with chronic hepatitis C, parameters for the prediction of SVR are important to be able to estimate the potential for treatment success (3). Genetic variations, apart from environmental and viral factors, are probably involved in the efficacy of interferon-based therapies. The IL28B gene encodes cytokine IL28B (IFN λ3). It belongs to the type 3 IFN family, which also includes IL-29 (IFN λ 1) and IL-28A (IFN λ2). This cytokine plays a critical role in clearing viral infections through inhibition of virus replication (11). Several studies have reported associations of different SNPs in an IL28B gene region with response to antiviral therapy (5, 12-14). Chu et al. (15) showed that the determination of IL28B polymorphism may be important for the analysis of treatment outcomes in clinical trials of interferon-free regimens.
In this study, we determined rs12979860 polymorphism in the IL28B gene in 45 responders and in 30 nonresponders under antiviral therapy. Our results showed that in the nonresponder patients, 6.7% had the CC genotype, 43.3% the CT genotype and 50% the TT genotype compared to the respondent patients in which the proportion of rs12979860 CC was 28.9%, CT was 37.8% and TT was 33.3%. In a study by Asselah et al. (16) on European and African populations, the frequency of the CC genotype was 26.8%, of the CT was 52.4% and of TT was 20.8%. In another study conducted by Xie et al. (17) on a Chinese population, the frequencies of the CC, CT and TT genotypes were 71.4%, 25.0% and 3.6%, respectively, in the SVR group and were 15.8% CC, 60.5% CT and 23.7% TT in the group with null virological response. In another study, of Italian patients of Caucasian ethnicity affected by chronic hepatitis, the genotype frequencies in the controls and in the chronic hepatitis C patients were as follows: for CC, 47.0% vs. 32.6%, for CT, 41.8% vs. 52.8% and for TT, 11.2% vs. 14.6% (18). The difference between the frequencies of genotypes in different studies can be explained by race, the sample size, the sampling and the population studied.
In the main part of our study, we compared the allele and genotype frequencies of the IL28B rs12979860 polymorphism in the two groups. A significant difference was observed in allele and genotype frequencies between the responder and the nonresponder groups. According to our results, carriers with the CC genotype had a higher chance of achieving SVR compared with those with the CT or TT genotypes. Our findings are in agreement with Sarrazin et al. (3) and Akuta et al. (19), whose results showed that SVR in patients with a rapid virological response was associated with the rs12979860 CC genotype, while for non-SVR, no association was found. Asselah et al. (16), in 82 treated patients, determined that the genotype distributions for IL28B polymorphism were significantly different between the responder and the nonresponder patients, and these researchers’ findings showed a better treatment response rate of the CC genotype of the IL28B gene SNP rs12979860. In Sharafi et al.’s (20) study, the differences in the distribution of IL28B rs12979860 genotypes between patients with chronic hepatitis C and healthy individuals were not statistically significant. In another study in Iran of Caucasian populations with hepatitis C infection, the results showed a higher SVR rate in patients with the CC genotype compared to those with the TT genotype; however, no significant difference was noted between the CT and TT genotypes. Also in agreement with our study, Daneshvar et al. (21) showed that patients with the C allele had a significantly higher SVR rate than did those with the T allele.
In the other part of the study, we investigated the association between blood HCV levels and IL28B rs12979860 polymorphism and did not find a significant association. This result disagrees with the results of some recent studies that have noticed a relationship between the responder allele and higher blood HCV levels. According to the results of Ge et al.’s (12) study, the CC genotype of rs12979860 was statistically associated with a higher baseline viral load. Another study has shown that in HCV-infected Caucasians, the rs12979860 genotype was associated with response to treatment, with the greatest effect seen when comparing subjects with the CC genotype to those with CT or TT. The presence of the CC genotype conferred nearly six fold increased odds of SVR relative to the CT/TT genotype (22). One explanation of this controversy may be low sample size.
All HCV-infected patients had either genotype 1 or 3. We determined the association between the different genotypes and IL28B rs12979860 polymorphism and found no significant association between the genotypes and the polymorphism. This was in contrast to the study of Falleti et al. (18), in which the CC genotype frequency was higher in HCV genotype 3-infected patients in comparison to those infected with HCV genotype 1; conversely, they found that the presence of the TT genotype was significantly associated with HCV infection with genotype 1 as compared to genotype 3. According to these researchers’ results, in chronic hepatitis C, the frequency of HCV infection due to unfavorable HCV genotypes 1, 4 and 5 increased from the IL28B rs12979860 CC genotype to the CT and TT genotypes. The researchers compared the genotype frequencies of this polymorphism between samples infected with HCV 1, 2 and 3 and showed that the CC genotype was most common in genotype 3 patients, followed by genotype 2 and then genotype 1. This controversy might be due to the larger number of genotype 3 patients and the absence of genotypes 2 and 4 in our study.
We did not find any relationship between viral load and virus genotypes in our study population, in agreement with Hadinedoushan et al.’s (23) study, which also failed to show any difference between HCV viral load and genotypes 1 and 3. Our study indicates that patients with the homozygous CC genotype had a significantly higher rate of response to treatment than did those with the TT or CT genotypes. Also, the IL28B rs12979860 polymorphism does not affect the blood viral load.