1. Background
Human cytomegalovirus (HCMV) infections are a major cause of morbidity and mortality among immunocompromised patients. Susceptible groups include renal transplant recipients, bone marrow recipients, and AIDS patients (1, 2). Hence, early diagnosis for the accurate treatment of HCMV is especially necessary to avoid poor clinical outcomes (3-6). Antiviral agents are used in the management of HCMV infections. Currently, the most common antiviral agents include ganciclovir (GCV), foscarnet, and cidofovir. These agents inhibit HCMV DNA synthesis by suppressing the DNA polymerase of the causative virus. Human cytomegalovirus infections usually require long-term treatment with antiviral agents for efficacy against the infective viruses. This can give rise to resistant strains of HCMV (7-10). Resistance to GCV has been evaluated by many researchers because this agent is used as the first-line treatment in 90% of HCMV patients (11).
Ganciclovir, a nucleoside analogue, must be phosphorylated to exert antiviral activity as an inhibitor of viral DNA polymerase UL54. The UL97 kinase, a virus-encoded product, activates the drug by monophosphorylation (12-17). HCMV resistance to GCV is mainly associated with the presence of mutations within UL97, DNA polymerase, or both viral genes (12, 18-25). Although mutations conferring GCV resistance have been mapped in both genes, the majority of the mutations appear in the UL97 gene, rather than as UL54 gene mutations (26). In the last two decades, several methods based on phenotypic testing have been described for the detection of HCMV-resistant mutations, such as the plaque reduction assay, ELISA, and DNA hybridization assays (27-29). However, these tests are burdensome, time-consuming, and labor-intensive (30). Therefore, there is an increasing need for early detection of the emerging mutants (31, 32).
2. Objectives
The purpose of the present study was the examination of GCV-resistant HCMV in Iranian immunosuppressed patients, before and after treatment.
3. Patients and Methods
3.1. Patients
A total of 87 specimens from Iranian patients were targeted in the present study. Clinical isolates from 48 bone marrow transplant patients and 39 renal transplant patients with suspected HCMV infections were identified and sent to the Keyvan virology specialty laboratory (KVSL) for molecular examination of the UL97 gene. The specimens were collected at two different stages of the disease: early (before GCV was initiated) and late (after six months of GCV therapy). All of the selected patients completed questionnaires and informed consent forms for participation in this study.
3.2. Nucleic Acid Extraction
Plasma samples (200 µL) were used for DNA extraction using the QIAamp DNA blood mini kit (Qiagen, USA). The extractions were carried out according to the manufacturer’s instructions.
3.3. Detection of HCMV in Specimens
Extracted DNA of plasma samples was subjected to TaqMan® real-time PCR targeting the UL83 gene, with the primers and probe proposed by Khansarinejad et al. (33). The sequence of the forward primer was 5′-TGAACATCCCCAGCATCAACG-3′, of the reverse primer was 5′-CAGTCCCGAGACMGTGAGAC-3′, and of the probe was 5′-FAM TGCCACATCTGCTTGCCCGACGC-BHQ1-3´. These primers and probe amplified the 137 bp segment of the UL83 gene in HCMV. Internal control of DNA extraction and PCR amplification was conducted by amplification of the human β globin gene. The primers consisted of the following sequences: forward primer: 5′-CAGAGCCATCTATTGCTTAC-3′, reverse primer: 5′-CATGGTGTCTGTTTGAGGTT-3′, and probe: 5′-JOE-ACACAACTGTGTTCACTAGC-BHQ1-3′ (34). All primers and probes were purchased from Bioneer Co., Ltd. (Daejeon, Korea). Real-time PCR was performed with TaqMan universal master mix (Takara Bio, Shiga, Japan). DNA was added to a 25 PCR mixture containing 12.5 μL of TaqMan universal PCR master mix, 300 nM of each primer, and 200 nM of probe. The reaction consisted of 10 minutes at 95°C, followed by 40 cycles of amplification, including 15 s at 95°C and 1 minute at 60°C.
3.4. Analysis of the UL97 Gene
Specific regions of the UL97 gene of HCMV in the early and late samples were amplified using nested PCR, as defined by Benzi et al. (35). The first round was done with the following primers: UL97-F1 (5′-GTTTCATCACGACCAGTGGA-3′) and UL97-R1 (5′-GGTCCTCCTCGCAGATTATG-3′). The second round was carried out with UL97-F2 (5′-TCATCACGACCAGTGGAAGCT-3′) and UL97-R2 (5′-GCGACACGAGGACATCTTGG-3′). The UL97 fragment was amplified using 1.5 mM MgCl2, 0.2 mM dNTP, 1 U Taq DNA polymerase, 10X PCR buffer (Kowsar Biotech Co., Tehran, Iran), 300 nM of each primer (Bioneer Co., Daejeon, Korea), and 50 ng genomic DNA of HCMV. The thermal cycler was run at 94°C for 30 seconds, 52°C for 40 seconds, and 72°C for 2 minutes for 40 cycles, and 72°C for 7 minutes. All of the nested PCR products were subjected to direct Sanger dideoxy DNA sequencing using internal primers of the UL97 gene (UL97-F2 and UL97-R2) (Bioneer Co., Daejeon, Korea). The results of the sequencing were analyzed with the CLC sequence viewer, version 6.4 (CLC Bio Co., Aarhus, Denmark).
4. Results
The fluorescent signals were observed in the FAM and JOE channels related to the UL83 and β globin genes, respectively. These data showed specific amplification of UL83 and β globin genes by real-time PCR amplification of genomic DNA of all samples. To confirm the specificity of the PCR and the real-time PCR assay, genomic DNA from other human herpesviruses (herpes simplex virus types 1 and 2, varicella-zoster virus, and human herpesvirus 6) was amplified. The negative results after amplification of all samples proved the specificity, and there was no cross-reactivity of all assays (data not shown).
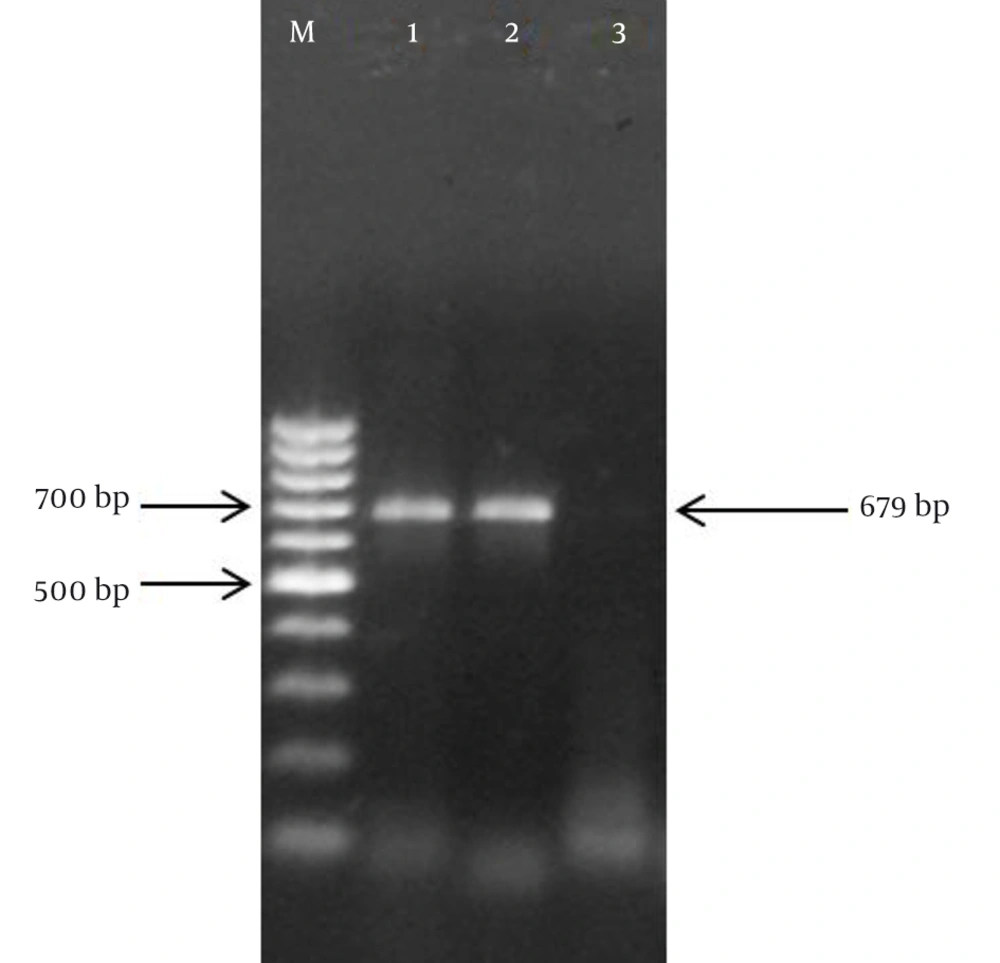
Specific nested PCR primers amplified the vulnerable region in the UL97 gene. The binding site of primers on the UL97 gene of HCMV (2124 bp) were nucleotide numbers 1254 - 1273, 2070 - 2089, 1257 - 1277, and 1916 - 1935 for UL97-F1, UL97-R1, UL97-F2, UL97-R2, respectively. Accordingly, in the first round of the assay, the expected 836-bp PCR products were exhibited. The presence of the expected 679-bp band in the second round of the assay proved the specific amplification of the UL97 gene during the nested PCR reaction (Figure 1).
Based on the previous report, the predominant codon numbers in the UL97 gene of HCMV corresponding to GCV resistance have been reported: L405P, M460I, M460T, M460V, V466G, H520Q, ACRA591-594Del4, 591-607Del, C592G, A594E, A594G, A594P, A594T, A594V, L595F, L595S, L595W, L595Del, 595-603Del, E596G, G598S, K599T, L600Del, T601Del, 601-603Del, C603R, C603S, C603W, C607F, and C607Y (36). In order to analyze the sequenced PCR product of the UL97 gene, a series accession number from the GenBank database was selected. Then, using the CLC Sequence Viewer software, the unknown sequences of the UL97 gene from Iranian immunosuppressed patients were aligned with wild-type and mutant nucleotide sequences.
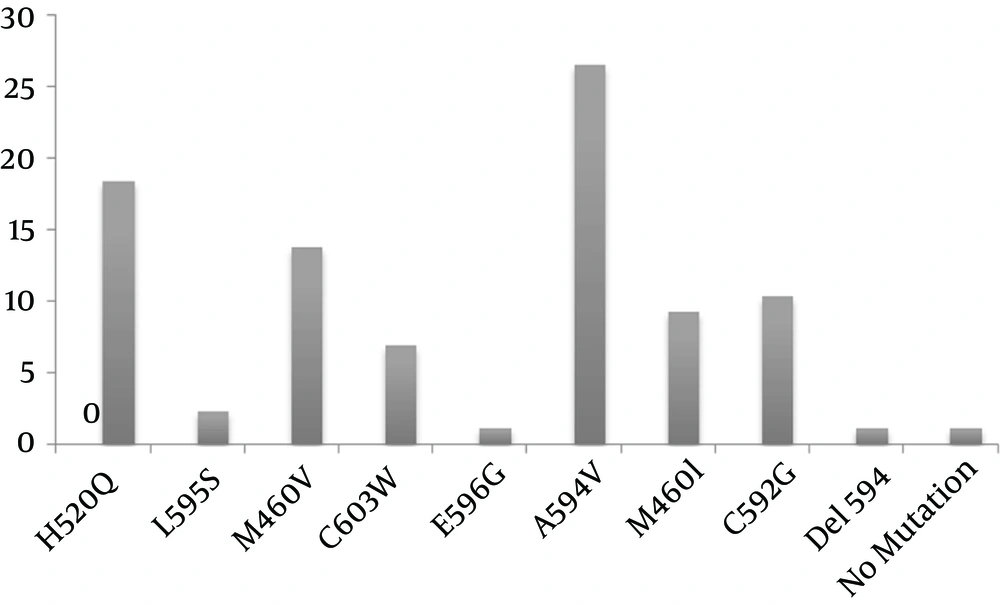
The codon locations of the samples were evaluated with the reported mutant codon, which has been published previously (12, 31, 35, 36). Of the 87 samples, only one (1.14%) point mutation occurred in the early-phase analysis of the samples, but many mutations occurred after treatment with GCV: A594V (26.46%), H520Q (18.39%), M460V (13.79%), C592G (10.34%), M460I (9.19%), C603W (6.89%), L595S (2.29%), E596G (1.14%), and Del 594 (1.14%) (Figure 2). Comparison of the early and late sequences of the UL97 gene exhibited the presence of point mutations in the specific codon of the UL97 gene mainly after drug consumption in the late phase. In addition, for the first time among immunocompromised patients in Iran, a new mutation (Del codon 594) was discovered in the HCMV UL97 gene in a patient who had undergone renal transplantation. The frequencies of early and late mutations of the UL97 gene are listed in Table 1. The mutant sequence has been submitted to GenBank (KM081676).
| CN | WTN | MN | WTAN | MAN | EAM | LAM |
|---|---|---|---|---|---|---|
| H520Q | CAT, CAC | CAA, CAG | AY642448 | AY642438 | 0 | 16 |
| L595S | CTT, CTC, CTA, CTG | TCT, TCC, TCA, TCG | HQ158765 | AY642437 | 0 | 2 |
| M460V | ATG | GTT, GTC, GTA, GTG | AY642435 | HCU07356 | 0 | 12 |
| C603W | TGT, TGC | TGG | AY642443 | AY642451 | 0 | 6 |
| E596G | GAA, GAG | GGT, GGC, GGA, GGG | AF345351 | - | 0 | 1 |
| A594V | GCT, GCC, GCA, GCG | GTT, GTC, GTA, GTG | AY642439 | HM208324 | 1 | 23 |
| M460I | ATG | ATT, ATC, ATA | HQ158775 | AY642447 | 0 | 8 |
| C592G | TGT, TGC | GGT, GGC, GGA, GGG | HQ158791 | HM352648 | 0 | 9 |
| Del 594 | GCG | - | KC664775 | KM081676 | 0 | 1 |
| No mutation | - | - | BK000394 | - | 0 | 1 |
List of Accession Numbers (Wild-Type and Mutant Sequences) and Frequency of Early and Late Mutations Related to the UL97 Gene
5. Discussion
Prolonged GCV therapy in transplant patients is the main cause of mutations occurring in the UL97 gene and the subsequent emergence of GCV resistance. One of the most difficult therapeutic challenges is to determine the wild-type or mutant viruses in transplant patients (37, 38). Mutations of M460V, M460I, H520Q, A594V, L595S, L595F, C603W, and C607Y for resistance of HCMV to GCV have already been reported. In addition, N510S, A590T, A591V, A591D, C592G, L595W, L595T, E596G, E596D, N597I, G598V, K599M, C603Y, A606D, and V665I have also been discovered among clinically GCV-resistant isolates (39). Our study results showed that the early-phase analysis of mutations identified only one renal transplant patient’s sample as positive for the presence of A594V. This suggests that the presence of resistant viruses is weak in the early stages of the disease, whereas a high frequency of the mutated viruses can be detected in the late analysis of mutations.
Erice et al. elucidated no evidence so far for GCV resistance of A591V, C592G, L595W, L595T, E596G, N597I, G598V, K599M, C603Y, A606D, and V665I. We found one point mutation of E596G among our clinical specimens with resistance to HCMV (39). Hantz et al. evaluated 18 infected patients, and discovered high-level resistance mutations of M460V (3/17), A594V (2/17), L595S (6/17), and L595F (2/17), and a 12-codon deletion (Del 591–603). They also found low-level resistance mutations of C592G, A594P, and C603W in the UL97 gene (11). Our data also support the fact that most of the mutations are seen in the UL97 gene, with a high frequency in the A594V (23/87), H520Q (16/87), and M460V (12/87) codons.
Some deletions have previously been explained in an immunocompromised patient in association with clinical resistance to GCV, in the codons of the UL97 gene. Hantz et al. discovered a novel GCV-resistance mutation (a deletion of codon 601 of the UL97 gene) in kidney transplant patients (12). Marfori et al. found a new viral UL97 mutation (deletion of codons 601 - 603) in stem cell transplant recipients (40). Gentry et al. discovered that a deletion in base pair 498 of UL97 is responsible for drug resistance in the HCMV virus (41). Chou et al. reported a deletion mutation (codons 595 to 603) in the UL97 gene (16). For the first time, our study presents a description of a UL97 codon 594 deletion in a GCV-resistant HCMV isolate among Iranian immunocompromised patients. This deletion emerged in a renal transplant recipient after six months of GCV treatment for HCMV infection.
5.1. Conclusion
The results of our study support the hypothesis of a high prevalence of A594V, M460V, and H520Q mutations, low rates of point mutations in the L595S and E596G codons, and an intermediate prevalence of the C603W, M460I, and C592G mutations. Furthermore, the presence of Del 594 in Iranian immunosuppressed patients was also observed. The present study demonstrates an effective method for the early detection of mutations that cause GCV resistance, thereby guiding the treatment of HCMV infections.