1. Background
The resistance of major gram-negative bacteria, such as Escherichia coli and Klebsiella, against antibiotics of the beta-lactam group, especially carbapenem, is increasing. These bacteria isolated from infections in hospitals all over the world result in the failure of treatment (1).
The combined use of aminoglycoside and beta-lactam group antibiotics increases permeability by chelation of bacterial cell walls. However, a high level of resistance against aminoglycoside is evident in strains that are resistant to beta-lactam group antibiotics (2, 3). This high resistance needs to be investigated in detail, and the molecular mechanisms and epidemiological features of aminoglycoside resistance should be the focus. According to molecular studies conducted to determine the resistance mechanisms in bacteria against aminoglycoside, the following three mechanisms are responsible for the resistance: enzymes that modify aminoglycoside, mutation in genes that transport aminoglycoside, and alteration in ribosomal target proteins (4). A new resistance mechanism was discovered in 2003. This mechanism suggests that 16S rRNA methyltransferase genes are transported with plasmids and leads to a high level of resistance to aminoglycoside. This mechanism was demonstrated in Pseudomonas aeruginosa isolates in Japan and in Enterobacteriaceae isolates in France (5, 6). 16S rRNA methyltransferase was reported to significantly increase the level of aminoglycoside resistance by methylation of the G1405 residue (minimum inhibitory concentration [MIC] is typically ≥ 256 µg/mL) (7).
16S rRNA methyltransferase enzymes are generally seen together with carbapenemases and extended-spectrum beta-lactamase (ESBL). The existence of 16S rRNA methyltransferase has been observed in isolates carrying the genes blaNDM-1, blaKPC-2, blaVEB, blaTEM, blaCTX-M, blaSHV, and blaOXA. This outcome can be attributed to beta-lactamase and methyltransferase enzymes being located on the same plasmid (8-11).
2. Objectives
This study aimed to detect the co-existence of 16S rRNA methyltransferase enzymes and beta-lactamase genes in multidrug resistant (MDR) Klebsiella pneumoniae strains isolated from clinical samples in healthcare-associated infections.
3. Methods
A total of 228 clinical samples (i.e., blood, urine, bronchoalveolar lavage, sputum, and surgical wound) were collected from different clinics of the Medical Faculty of Balcali hospital at Cukurova University, Turkey, between August and December 2014. All the laboratory tests were performed at the laboratory of the Center and Medical microbiology department of the university. The samples were cultured on blood agar and Endo agar plates (Merck, Germany) at 37°C for 24 hours. The isolates were first evaluated by Gram staining to examine the morphology of colonies and biochemical test characteristics. Then, the bacterial species were identified using the VITEK-2 automated identification system (Biomerieux, Basingstoke, UK).
The resistance of beta-lactam antibiotics was investigated by the Kirby-Bauer disk diffusion (KBDD) test. The presence of ESBL was verified by the double disc synergy test, and carbapenem resistance was determined using the modified Hodge test. Escherichia coli ATCC 25922 was used as the control culture. To detect aminoglycoside resistance, amikacin (30 µg) and gentamicin (10 µg) sensitivities were examined through the KBDD test. Minimum inhibitory concentrations were determined by the agar dilution method on Mueller-Hinton agar plates according to the protocol recommended by the European committee on antimicrobial susceptibility testing (12).
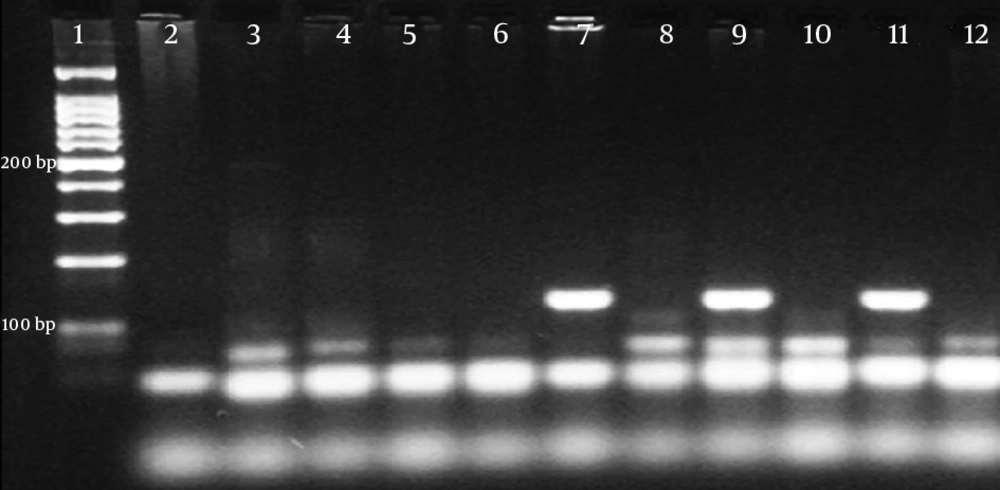
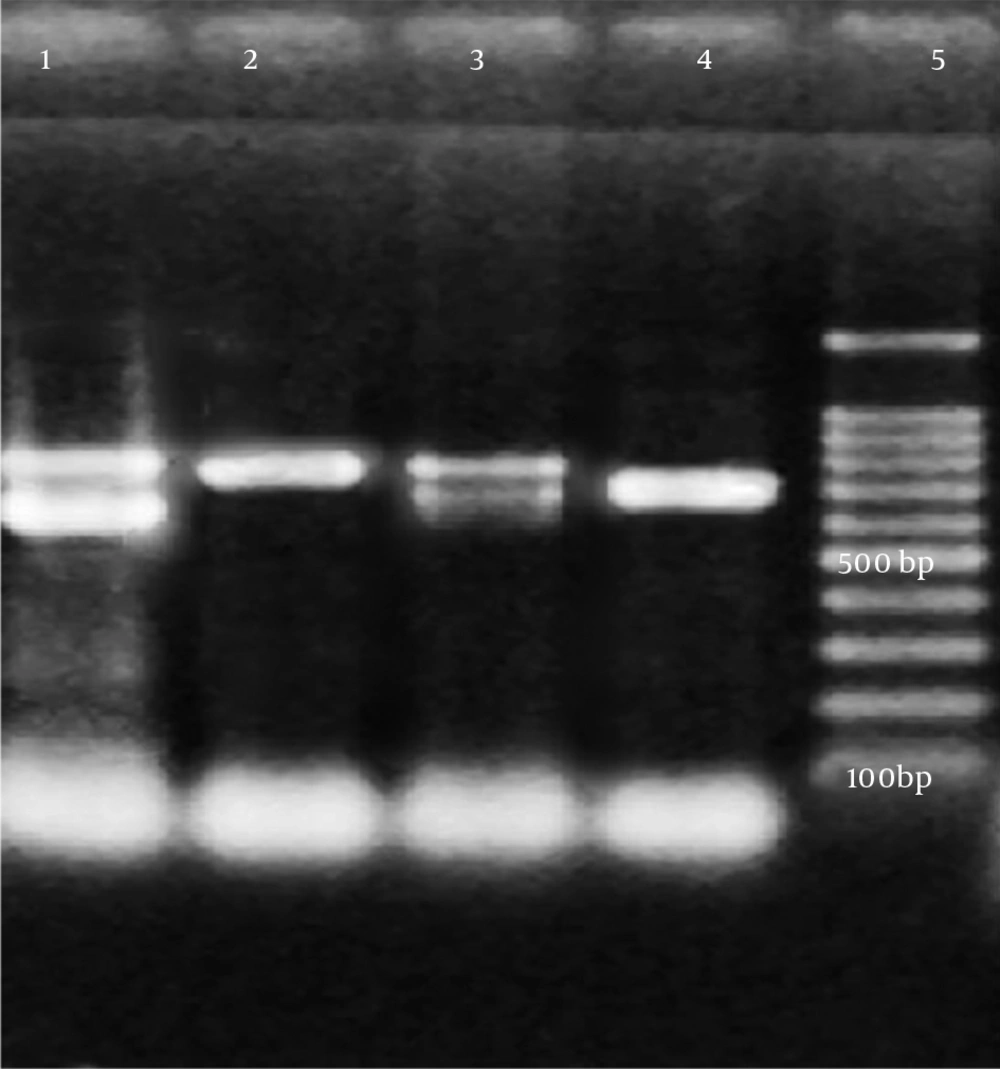
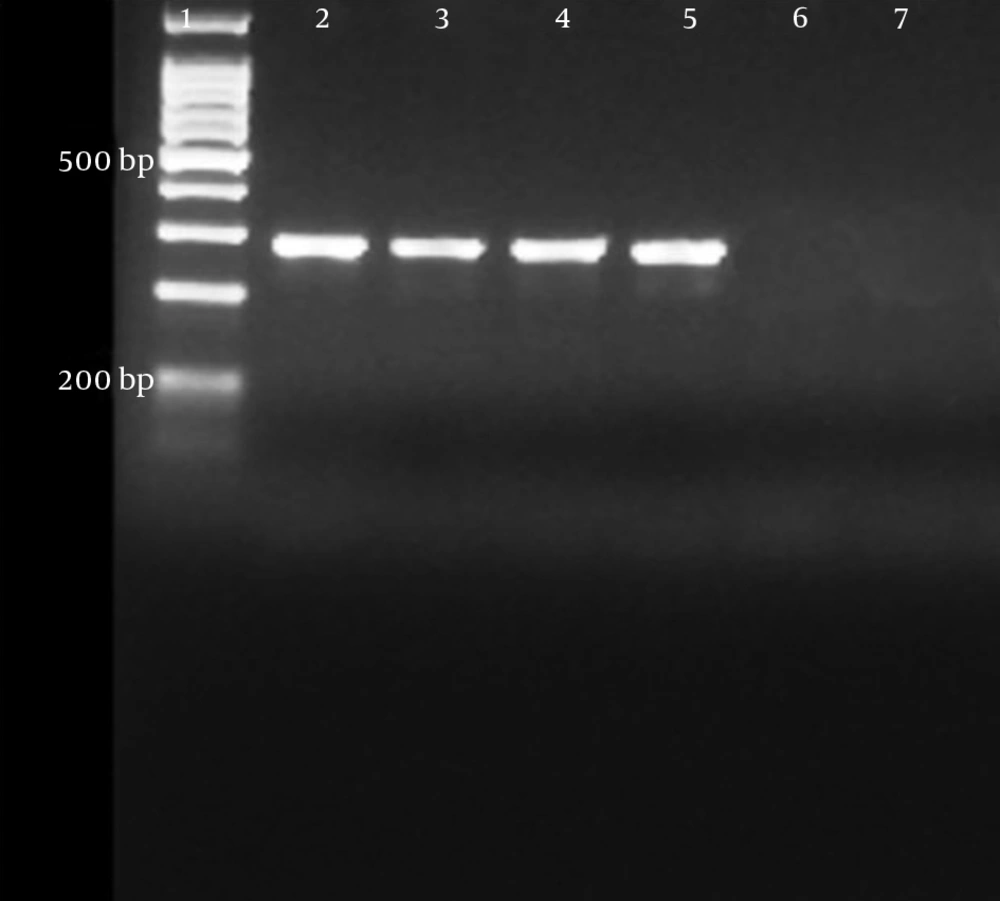
In all isolates, DNA extraction was performed mechanically with a Mickle cell disruptor (The Mickle Lab. Engineering Co. Ltd., Gamshall, Surrey, UK). However, polymerase chain reaction (PCR) was performed only in isolates with an MIC value of > 512 μg/mL for amikacin and > 128 μg/mL for gentamicin to detect beta-lactamase genes and 16s rRNA methyltransferase genes. The multiplex PCR method was used to determine the presence of armA, npmA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, and rmtH genes (13). In addition, a type-specific multiplex PCR protocol was used to determine the carbapenemase and ESBL genes (Table 1) (14-16). The PCR products were separated on 1.5% agarose gel and monitored using a Kodak UV transilluminator (Kodak, New York, USA). A sequence analysis was performed using the dye terminator cycle sequencing method and the ABI Prism Big dye terminator kit (Applied Biosystems, Foster City, USA). The assay was conducted according to the manufacturer’s standard protocol. Data were collected on an ABI 310 automated fluorescence sequencer (Applied Biosystems). The types of 16S rRNA methyltransferase, carbapenemase, and ESBL genes were identified by comparing them with sequences from the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). In all PCR procedures, distilled sterile water was used as the negative control. As no reference strains carrying rmtC were available, a positive control could not be used. Nevertheless, the sequences were verified by the DNA sequencing method.
| Multiplex Primers | Gene | Forward/Reverse | Sequences (5’-3’) | Product Size |
|---|---|---|---|---|
| TEM, SHV and OXA-1-like | TEM | F | CATTTCCGTGTCGCCCTTATTC | 800 bp |
| R | CGTTCATCCATAGTTGCCTGAC | |||
| SHV | F | AGCCGCTTGAGCAAATTAAAC | 713 bp | |
| R | ATCCCGCAGATAAATCACCAC | |||
| OXA-1 like | F | GGCACCAGATTCAACTTTCAAG | 564 bp | |
| R | GACCCCAAGTTTCCTGTAAGTG | |||
| CTX-M group 1, group 2 and group 9 | CTX-M group 1 | F | TTAGGAARTGTGCCGCTGYAb | 688 bp |
| R | CGATATCGTTGGTGGTRCCATb | |||
| CTX-M group 2 | F | CGTTAACGGCACGATGAC | 404 bp | |
| R | CGATATCGTTGGTGGTRCCATb | |||
| CTX-M group 9 | F | TCAAGCCTGCCGATCTGGT | 561 bp | |
| R | TGATTCTCGCCGCTGAAG | |||
| CTX-M group 8/25 | CTX-M-8, 25,26,39,40,41 | F | AACRCRCAGACGCTCTACb | 326 bp |
| R | TCGAGCCGGAASGTGTYATb | |||
| VEB, PER and GES | GES | F | AGTCGGCTAGACCGGAAAG | 399 bp |
| R | TTTGTCCGTGCTCAGGAT | |||
| PER | F | GCTCCGATAATGAAAGCGT | 520 bp | |
| R | TTCGGCTTGACTCGGCTGA | |||
| VEB | F | CATTTCCCGATGCAAAGCGT | 648 bp | |
| R | CGAAGTTTCTTTGGACTCTG | |||
| SME,IMI | SME | F | GAGGAAGACTTTGATGGGAGGAT | 334 bp |
| R | TCCCCTCAGGACCGCCAAG | |||
| IMI | F | GGTGTCTACGCTTTAGACACTGGCTC | 536 bp | |
| R | GCACGAATACGCGCTGCACCGG | |||
| OXA-48-like | OXA-48-like | F | GCTTGATCGCCCTCGATT | 281 bp |
| R | GATTTGCTCCGTGGCCGAAA | |||
| IMP, VIM and KPC | IMP | F | TTGACACTCCATTTACDGb | 139 bp |
| R | GATYGAGAATTAAGCCACYCTb | |||
| VIM | F | GATGGTGTTTGGTCGCATA | 390 bp | |
| R | CGAATGCGCAGCACCAG | |||
| KPC | F | CATTCAAGGGCTTTCTTGCTGC | 538 bp | |
| R | ACGACGGCATAGTCATTTGC | |||
| GIM-1,SIM-1, NDM-1 and SPM-1 | GIM | F | CGTTGCCAGCTTTAGCTCAGG | 279 bp |
| R | GCAACTTGATACCAGCAGTGCG | |||
| SIM | F | TTGCGGAAGAAGCCCAGCCAG | 613 bp | |
| R | GCG TCT CCG ATT TCA CTG TGG C | |||
| NDM | F | CCC GGC CAC ACC AGT GAC A | 129 bp | |
| R | GTA GTG CTC AGT GTC GGC AT | |||
| SPM | F | GGG TGG CTA AGA CTA TGA AGC C | 447 bp | |
| R | GCC GCC GAG CTG AAT CGG | |||
| rmtA, rmtC, rmtD, rmtG, rmtH | rmtA | F | AAACTATTCCGCATGGTTC | 88 bp |
| R | TCATGTACACAAGCTCTTTCC | |||
| rmtC | F | CAGGGGTTCCAACAAGT | 246 bp | |
| R | AGAGTATATAGCTTGAACATAAGTAGA | |||
| rmtD | F | TCGTTTCAGCACGTAAAACA | 652 bp | |
| R | CAGCGCGAAATTCAAAAAGG | |||
| rmtG | F | ACGGAATGCCGCGCGAAGTA | 381 bp | |
| R | TCTCCGCAAGCAGATCGCCG | |||
| rmtH | F | ATGACCATTGAACAGGCAGC | 464 bp | |
| R | AGGGCAAAGGTAAAATCCCA | |||
| armA, npmA, rmtB, rmtE, rmtF | armA | F | ATTTTAGATTTTGGTTGTGGC | 101 bp |
| R | ATCTCAGCTCTATCAATATCG | |||
| npmA | F | GGGCTATCTAATGTGGTG | 229 bp | |
| R | TTTTTATTTCCGCTTCTTCGT | |||
| rmtB | F | ACTTTTACAATCCCTCAATAC | 171 bp | |
| R | AAGTATATAAGTTCTGTTCCG | |||
| rmtE | F | GATGCCGTGTCTGTTACGCCG | 446 bp | |
| R | ACGTGAACCCACGAGTCCTGC | |||
| rmtF | F | CGATCCTACTGGGCTCCAT | 314 bp | |
| R | GGCATAGTGCTTTTCCATGC |
4. Results
Forty isolates of K. pneumoniae were evaluated to determine the existence of and the relationship among carbapenemase, ESBL, and 16S rRNA methyltransferase genes. The strains were isolated from 4 blood, 15 urine, 7 bronchoalveolar lavage, 12 sputum, and 2 surgical wound samples. Twenty K. pneumoniae strains were resistant to amikacin, and 40 were resistant to gentamicin. The MIC values of amikacin and gentamicin were found to be > 512 µg/mL in five isolates and > 128 µg/mL in 10 isolates, respectively (Table 2).
| Amikacin MIC Value, µg/mL | Number of Isolates | Gentamicin MIC Value, µg/mL | Number of Isolates |
|---|---|---|---|
| > 512 | 5 | > 128 | 10 |
| 128 | 5 | 128 | 10 |
| 64 | 5 | 64 | 4 |
| 32 | 1 | 32 | 8 |
| 16 | 4 | 16 | 2 |
| - | - | 8 | 2 |
| - | - | 4 | 4 |
| Total | 20 | Total | 40 |
PCR was performed only on five isolates with an MIC value of > 512 μg/mL for amikacin and > 128 μg/mL for gentamicin. A specific DNA fragment with a length of 246 bp was amplified on the rmtC gene in four isolates with the above-mentioned MIC values. Of these four isolates, three contained the blaNDM-1gene and all were found to have at least one ESBL gene (Table 3). A sequence analysis was performed to confirm the PCR results.
| Isolates | Amikacin MIC value, µg/mL | Gentamicin MIC Value, µg/mL | 16S Rrna Methyltransferase Gene | Carbapenemase Genes | ESBL Genes |
|---|---|---|---|---|---|
| 1 (Urine) | > 512 | > 128 | rmtC | NDM-1 | SHV |
| 2 (Urine) | > 512 | > 128 | rmtC | NDM-1, OXA-48 | TEM,SHV |
| 3 (Blood) | > 512 | > 128 | rmtC | OXA-48 | TEM |
| 4 (BAL) | > 512 | > 128 | rmtC | NDM-1,OXA-48 | TEM,SHV |
| 5 (BAL) | > 512 | > 128 | - | OXA-48 | SHV |
5. Discussion
Combination antibiotic therapy has been shown to have synergistic effects in vitro and in vivo. This treatment alternative is usually preferred in clinics to prevent the development of resistance without requiring a toxic dose or widening the antibacterial spectrum (17). In particular, carbapenem and aminoglycoside are used in combination prior to colistin and tigecycline in the treatment of infections of Enterobacteriaceae isolates that produce carbapenemase. For example, the combinations of meropenem/imipenem and amikacin or doripenem and gentamicin have been suggested as an effective alternative treatment for K. pneumoniae strains that produce carbapenemase (18, 19).
In recent years, plasmids that transport ESBL and carbapenemase enzymes have been found to carry 16S rRNA methyltransferase aminoglycoside-resistant genes. Thus, aminoglycoside and beta-lactam antibiotic combinations are no longer considered clinically effective. This finding was verified by Wu et al., who detected the armA methyltransferase gene on a plasmid that codes the K. pneumoniae carbapenemase enzyme (20).
Despite having low prevalence in different types of bacteria, the 16S rRNA methyltransferase gene is globally spreading because of the plasmids that transport carbapenemase and ESBL genes among gram-negative bacilli. In Taiwan, the rmtB and armA genes were identified in E. coli and K. pneumoniae isolates with a high level of amikacin resistance, and the prevalence of armA was found to be higher than that of rmtB (7). In Japan, rmtA, rmtB, npmA, and armA were detected in gram-negative isolates with an amikacin MIC value of over 512 mg/L (21). In Korea, the co-existence of rmtE and CMY-2 cephalosporinase was reported (22). In China, armA and rmtB genes were found in gram-negative bacteria isolates with the amikacin MIC value being consistently over 512 mg/L (23).
In Saudi Arabia, the relationship between ESBL and 16S rRNA methyltransferase genes was investigated, and the rmtB, rmtC, and armA genes were detected in the same isolates (24). Fritsche et al. analyzed gram-negative bacteria isolates from North America, Latin America, and Europe and found that the armA, rmtB, and rmtD genes were resistant to aminoglycosides in isolates with amikacin MIC values of over 128 µg/L (25). The existence of the rmtF methyltransferase gene was reported in Nepal, the United States, India, and England (26). The rmtH gene was determined in a K. pneumoniae strain isolated from an American soldier who was wounded in Iraq in 2006 (27). In Turkey, the rmtB gene was shown in a K. pneumoniae isolate that was aminoglycoside resistant (28, 29). In another study, 16S rRNA methyltransferase resistance was not found in any of the 59 aminoglycoside-resistant clinical isolates that were evaluated over a five-year period (30).
In the current study, the rmtC gene was detected in four isolates with an MIC value of > 512 µg/mL for amikacin and > 128 µg/mL for gentamicin. We determined both NDM-1 and rmtC genes in three isolates with a high level of aminoglycoside resistance. We found at least one ESBL gene in all four isolates that contained the rmtC gene. The co-existence of the NDM-1 and rmtC genes was previously observed in K. pneumoniae strains isolated from clinical samples in Australia, Nepal, Kenya, and Bangladesh. However, to the best of our knowledge, this study is the first report conducted in Turkey showing the presence of the rmtC gene and the co-existence of NDM-1 and rmtC resistance genes among clinical isolates.
The combination of beta-lactam and aminoglycoside is an important treatment alternative for gram-negative bacterial infections, but it has lost its clinical significance because of 16S rRNA methyltransferase and other aminoglycoside-resistant mechanisms. 16S rRNA methyltransferase-type resistance has low prevalence but spreads easily in plasmids that transport beta-lactamase genes such as NDM-1. This resistance can spread across a wide geographic region in Turkey and in other parts of the world. Further studies are needed to determine the mechanisms of 16S rRNA methyltransferase and carbapenemase resistance. The spread of these types of resistances should be monitored using molecular analyses.