Dear Editor,
The Eimeria species are host-specific protozoan parasites that cause the disease known as coccidiosis in a variety of animals including cattle, sheep, goats, pigs and poultry throughout the world (1). These parasites invade epithelial tissues of the intestine, causing severe damage in the host and result in significant economic losses (2). At least twelve species of bovine Eimeria, ten species of ovine Eimeria and nine species of caprine Eimeria occur in Iran (3-5). Eimeria ahsata and E. crandallis are two of a group of ovine coccidia of what might be called the arloingi type (6). Conventionally, microscopic examination of Eimerian oocytes is the only practical method to discriminate species of Eimeria (2, 7, 8). However, the particular morphology of a given species may vary considerably. Accordingly, microscopic differentiation is not reliable because several species have confusing features, along with the presence of intraspecies variation (7). Therefore, morphological observations should not be used as isolated criterion for differentiation of species (9).
Thus, molecular assays have proven useful for the identification of Eimeria spp. to overcome the limitations of these traditional procedures (7, 10). However, hematological and biochemical changes in blood serum, and histopathological lesions of ovine coccidiosis in sheep and lambs naturally infected by E. crandallis and E. ahsata were mentioned in previous studies (11), but no information is available about the genetic characterization and phylogenic positions of E. crandallis and E. ahsata. In the present study, the 18S rRNA gene was employed as a molecular genetic approach to investigate the phylogenetic analysis and DNA sequence variations of E. crandallis and E. ahsata compared with other Eimeria that exist in the GenBank database. Therefore, the present study was undertaken to identify genetic characteristics of E. ahsata and E. crandallis in slaughterhouse wastewater samples.
Twenty-four grab samples of untreated wastewater (5 L each) were collected from the two slaughterhouse treatment plants located in the southwest suburban area of Tehran, Iran. The animals slaughtered in these two slaughterhouses included cattle, sheep and goats. Water samples were concentrated by the centrifugal (water-ether) concentration procedure, as previously described (12). Oocytes were sporulated in a 2% (w/v) potassium dichromate (K2Cr2O7) solution at room temperature (25°C) for three weeks. The sporulated oocytes were partially identified as E. ahsata and E. crandallis based on the morphology of oocytes at × 40 and × 1,000 magnifications (13). DNA was extracted using an AccuPrep® stool DNA extraction kit (Bioneer Corporation, Daejeon, South Korea) according to the manufacturer’s protocol. An approximately 636-bp fragment of DNA extract was PCR amplified on the basis of the 18S rRNA gene, as previously described (14). All PCR amplicons were sequenced in both directions on an automated DNA analyzer (ABI 3730 XL, Bioneer, South Korea).
Unique nucleotide sequences described in this work were deposited in GenBank and accession numbers KP893746, KP893747, KP893748 and KP893749 were provided for Iranian E. ahsata and E. crandallis sequences of 18S rRNA.
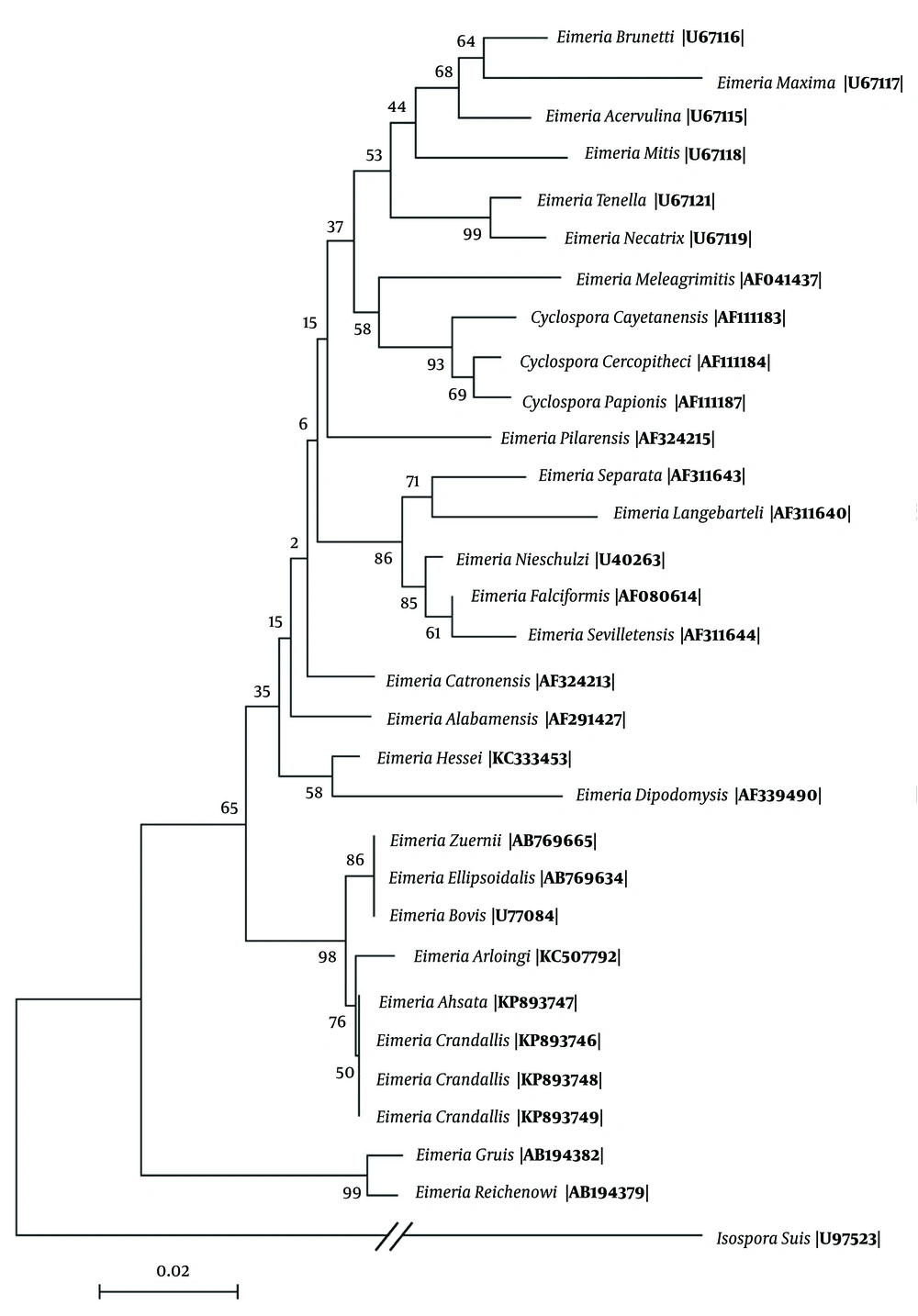
Sporulated oocytes of E. ahsata and E. crandallis were identified in all wastewater samples three weeks after their storage at 25°C. All samples that were found to be positive by microscopic evaluation were confirmed to be Eimeria by PCR. BLAST search of our 18S rRNA sequences against those previously published for other Eimeria spp. revealed the highest similarity (99% - 100% homology) with E. ahsata and E. crandallis, with differences observed at 2 - 4 nucleotides. Genetic distances among Eimeria species were in the range of 0.002 to 1.264 nucleotide substitutions per 100 bp. There was a minor difference in the sequence of 18S rRNA gene in E. ahsata and E. crandallis with E. arloingi available in GenBank. Phylogenetic analysis showed that one clade contained E. ahsata, E. crandallis and E. arloingi (the most pathogenic Eimeria in goats), and E. ahsata and E. crandallis grouped together in one clade (Figure 1).
Prior to this work, Eimeria parasites had not been reported in domestic wastewater samples. However, in the study conducted by Ben Ayed et al. (15) in Tunisia, E. alabamensis, E. ahsata and E. crandallis were found in urban wastewater, which was probably the result of effluents from cattle slaughterhouses in the city or fertilizers of animal origin used in parks and boulevards. In the present study, the microscopic examination resulted in positive results for all water samples from both slaughterhouse treatment plants. The presence of positive samples for E. ahsata and E. crandallis was confirmed by PCR assay.
In the present study, 18S rRNA sequences of E. crandallis and E. ahsata had considerable homology with E. arloingi, E. bovis and E. zuernii (98% - 99%). Similarly, Khodakaram-Tafti et al. (4) reported a close relationship between E. arloingi and bovine (E. bovis and E. zuernii) and ovine (E. ahsata and E. crandallis) coccidia, based on the 18S sequence analyses. It has also been shown that there is a similarity between these two species (E. crandallis and E. ahsata) and E. arloingi, based on microscopic evaluation of their sporulated oocytes. Identically, E. ahsata and E. crandallis in sheep, E. arloingi in goats and E. bovis and E. zuernii in cattle are highly pathogenic and formed a monophyletic group in the position away from other members in spite of many different biological characteristics and the histopathological lesions. The phylogram based on the 18S rRNA sequences showed that one clade contained E. ahsata, E. crandallis and E. arloingi, and E. ahsata and E. crandallis grouped together in one clade. Similarly, Bush (16) reported a tendency provided by the phylogenetic analysis of avian Eimeria for E. necatrix and E. tenella, the most pathogenic Eimeria in chicken.
While the 18S rRNA sequence may not eventually prove to be useful to examine in greater detail the magnitude of population variation of a single Eimeria species, it can be used to identify Eimerian oocytes in environmental samples.