1. Background
Staphylococcus aureus is one of the most important causative agents in both hospital- and community-acquired infections (1). This bacterium can cause various types of infection, including sepsis, pneumonia, wound sepsis, endocarditis, catheter-related infections, and urinary tract infections (UTIs) (2).
Methicillin-resistant Staphylococcus aureus (MRSA) strains are resistant to a large group of antibiotics called beta-lactams, including penicillins and cephalosporins (3). Almost all MRSA isolates have a penicillin-binding protein (PBP2a). PBP2a has much lower affinity to beta-lactam antibiotics compared to PBP2, which is the main physiological target of methicillin. PBP2a is encoded by the mecA gene (4).
Because of the high distribution of MRSA, these bacteria are now resistant to certain other antibiotics, such as tetracyclines, aminoglycosides, and lincosamides. These drug-resistant strains have become more difficult to treat (5, 6). Aminoglycosides are important antibiotics used to treat a variety of bacterial infections, particularly those caused by staphylococci (7). Aminoglycosides bind to the 30s ribosomal subunit of the bacterium and interrupt RNA translation, leading to bacterial death (8). This class of antibiotics are often used in combination with beta-lactams and glycopeptides to treat infections, such as bacterial endocarditis, caused by staphylococci and enterococci (9).
The three mechanisms of resistance to aminoglycosides are changes in the position of the ribosomal binding site for the drug, reduced permeability of the drug, and drug inactivation by enzymes. The enzymatic inactivation by aminoglycoside-modifying enzymes (AMEs) is an important mechanism of resistance in staphylococcal species. These enzymes are classified on the basis of modifying effects in three different categories: aminoglycoside acetyl transferases (AACs), aminoglycoside phosphotransferases (APHs), and aminoglycoside-nucleotidyltransferases (ANTs). Three enzymes, AAC (6’)/APH (2”), APH (3)-III, and ANT (4), are encoded by aac (6’)-Ie/aph (2”), aph (3)-IIIa, and ant (4)-Ia genes, respectively. These are the most common modifying enzymes among Staphylococcus species, and are clinically important (10).
High levels of resistance to methicillin, the continuing spread of MRSA strains, and the high resistance to aminoglycosides in these strains have been reported several times in Iran (11-13). With an emphasis on the role of combination therapy against staphylococcal infections, it is necessary to monitor the antibiotic susceptibility of S. aureus in order to identify and prevent the spread of resistant strains.
2. Objectives
The purpose of this study was to evaluate the drug-resistance profile of MRSA, as well as to detect aminoglycoside-resistant genes isolated from hospitalized patients in Sari and Tehran, Iran.
3. Methods
3.1. Bacterial Strains
In this cross-sectional study, 174 clinical isolates of MRSA were collected from specimens such as wounds, blood, sputum, tracheal exudate, bronchus, pleura, catheter, and urine of inpatients at three hospitals, Zare (Sari, Iran) and Loghman and Pars hospitals (Tehran, Iran), during March - November 2014. All isolates were cultured on mannitol salt agar medium (Merck, Germany), and identification of S. aureus was done using Gram staining and conventional biochemical tests, such as catalase, coagulase, and DNase (Merck, Germany) (14). Finally, the isolates were stored in tryptic soy broth (TSB) (Merck, Germany) containing 15% glycerol, and stored at -70°C until future processing.
3.2. Phenotypic Determination of MRSA
The disk diffusion method was used for the phenotypic detection of MRSA. For this purpose, oxacillin (OX) and cefoxitin (FOX) discs on Mueller-Hinton agar plates containing 4% NaCl were used, as recommended by the clinical and laboratory standards institute (CLSI) guidelines (15). Isolates with a zone of inhibition of < 21 for oxacillin and < 13 mm for cefoxitin were considered MRSA. Staphylococcus aureus ATCC700698 was used as the positive control for MRSA strains.
3.3. Antibiotic Susceptibility
To determine the antibiotic susceptibility pattern of MRSA strains, sensitivity to the following antibiotics was assessed: rifampin (5 μg), trimethoprim + sulfamethoxazote (30 μg), gentamycin (10 μg), erythromycin (15 μg), ciprofloxacin (5 μg), clindamycin (2 μg), cefazolin (30 μg), doxycycline (30 μg), and cefoxitin (30 μg) (Mast, UK). Antibiotic susceptibility testing was performed according to the Kirby-Bauer disk diffusion method and the results were interpreted with respect to CLSI guidelines (15). In addition, the minimum inhibitory concentration (MIC) of vancomycin was determined with the standardized micro-dilution broth method according to the CLSI.
3.4. DNA Extraction and Polymerase Chain Reaction
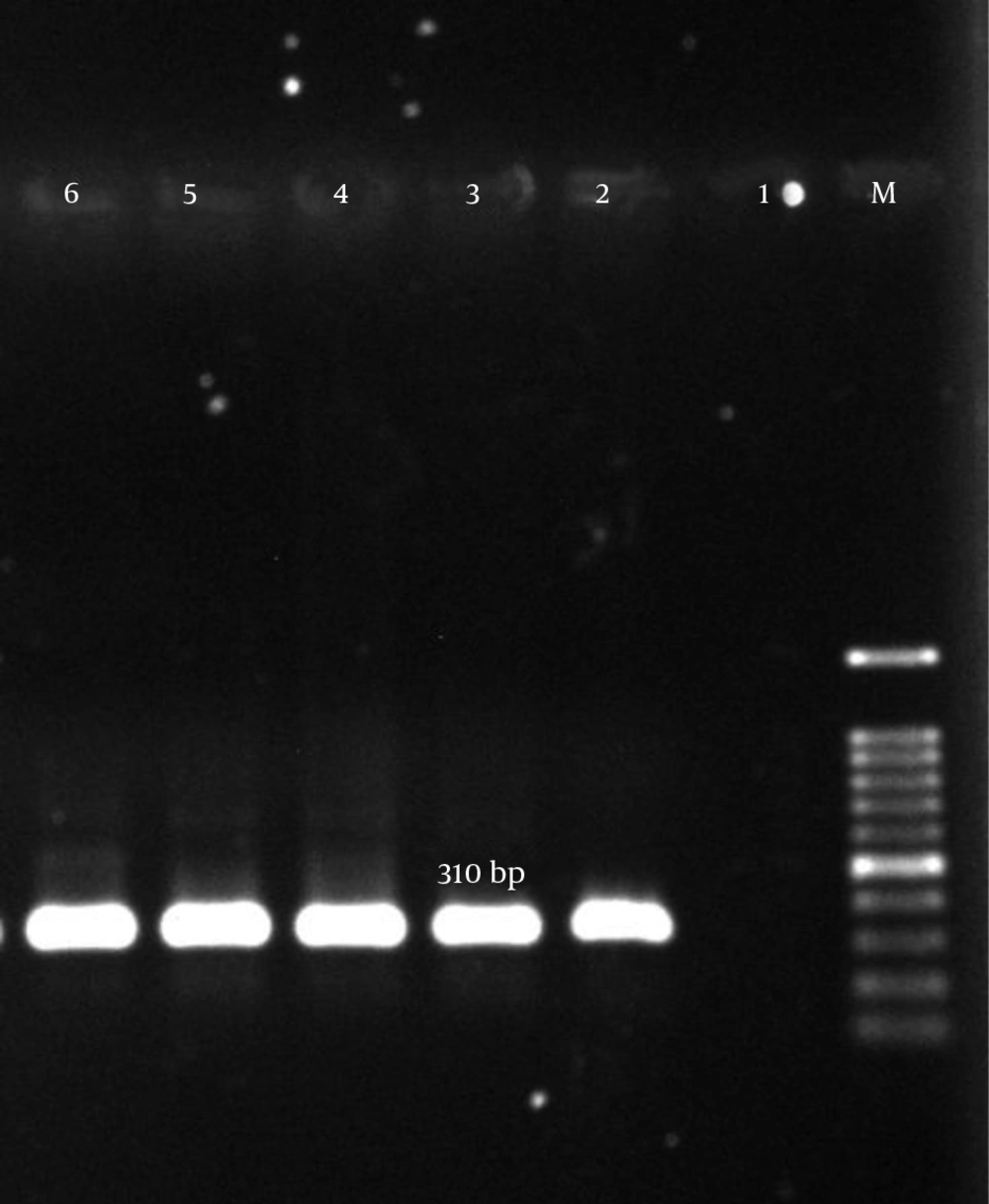
Genomic DNA was extracted using a commercially available DNA extraction kit (QiaAmp DNA Mini Kit) according to the manufacturer’s instructions. For confirmation of MRSA, the mecA gene was detected by PCR with the specific primers listed in Table 1 (16). Staphylococcus aureus ATCC700698 was used as the positive control for mecA gene detection (17).
| Gene | Primer Sequencimg | Size, bp | Reference |
|---|---|---|---|
| mecA | 5’GTAGAAATGACTGAACGTCCGATAA3’ | 310 | (4) |
| 5’CCAATTCCACATTGTTTCGGTCTAA3’ | |||
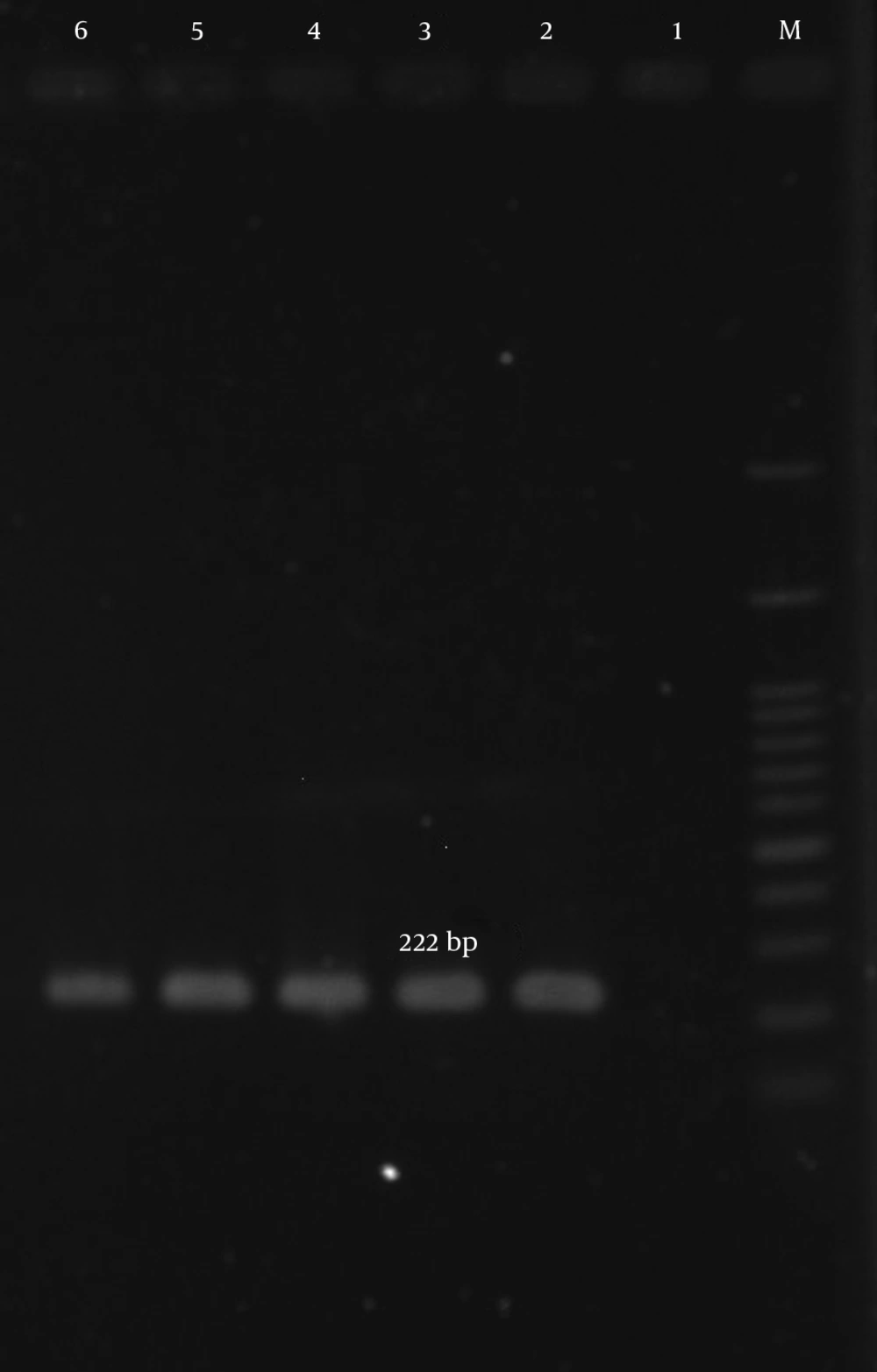
| aac(6’)-Ie/aph(2” ) | 5’CCAAGAGCAATAAGGGCATACC3’ | 222 | (18) |
| 5’CACACTATCATAACCACTACCG3’ | |||
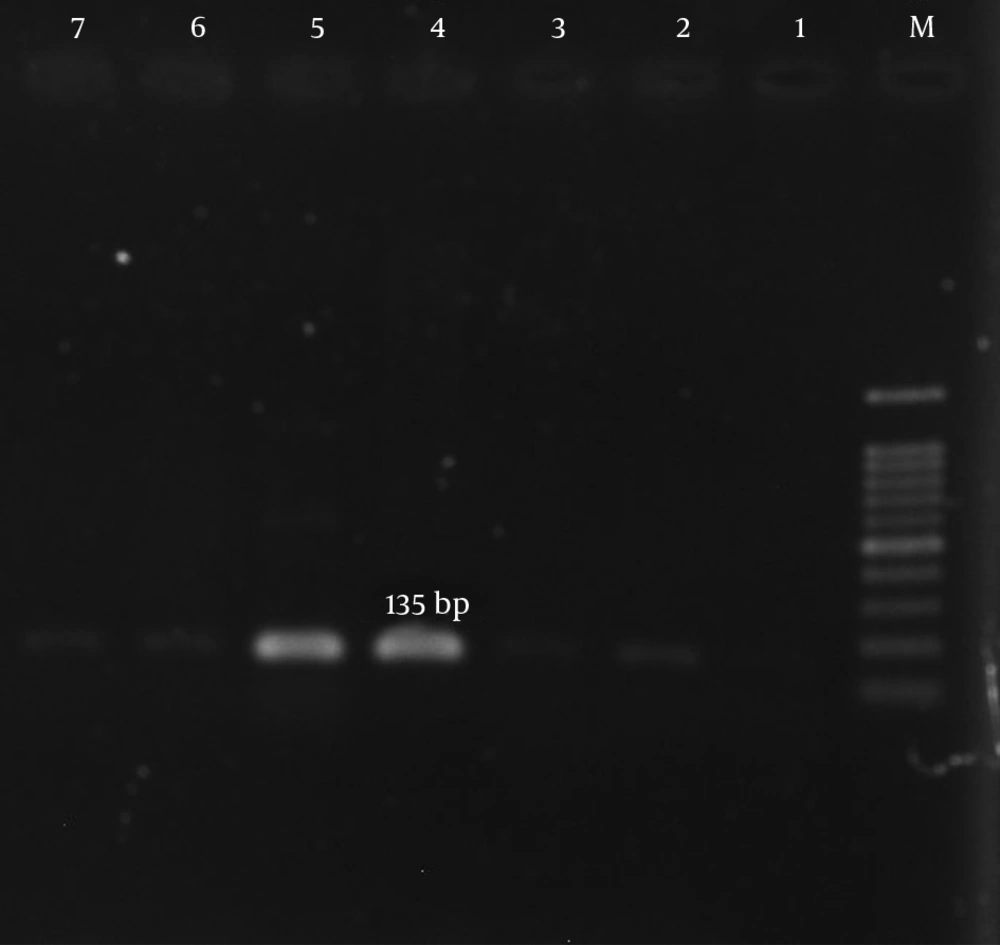
| ant(4’)-Ia | 5’AATCGGTAGAAGCCCAA3’ | 135 | (19) |
| 5’GCACCTGCCATTGCTA3’΄ | |||
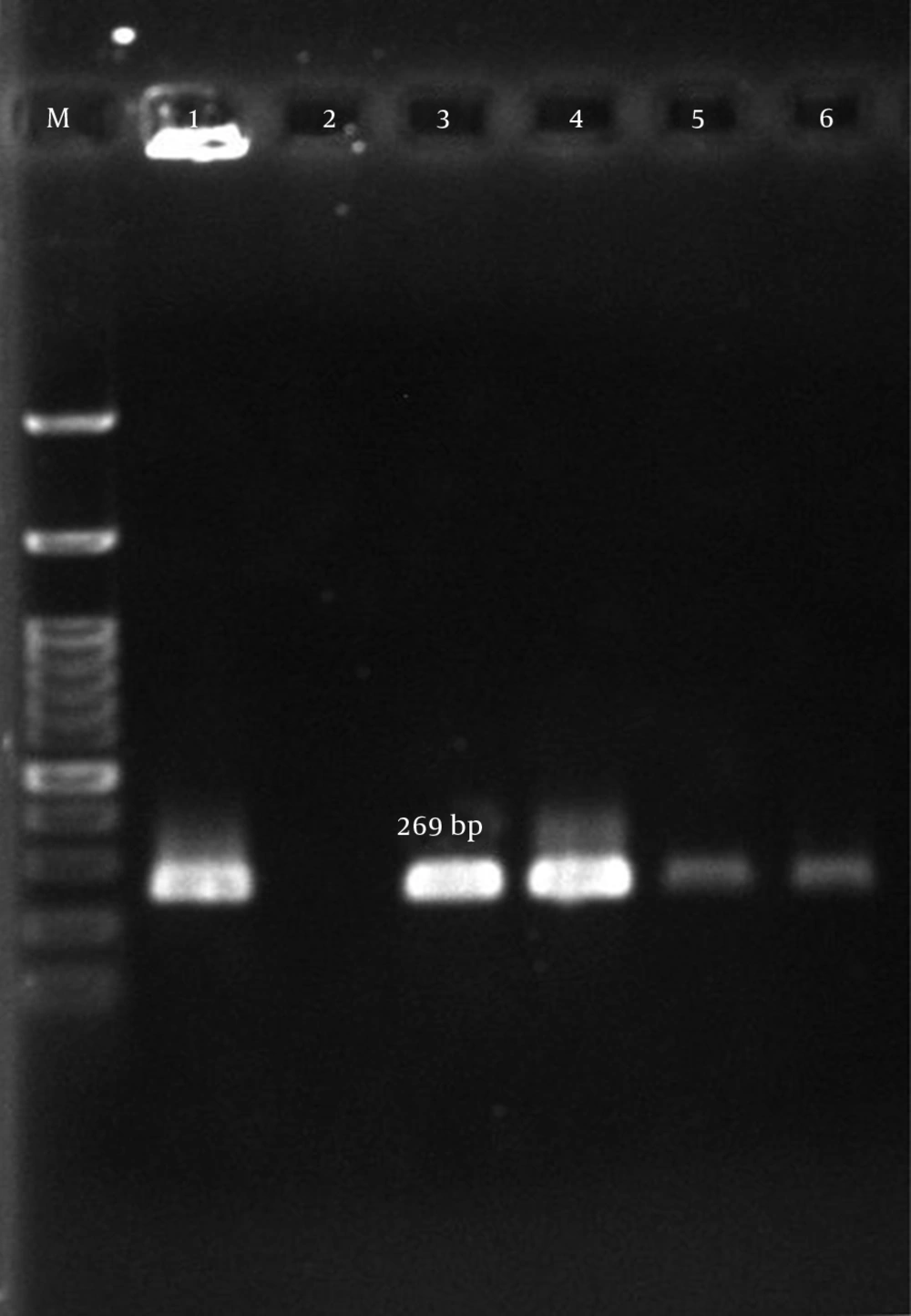
| aph(3’)-IIIa | 5΄CTGATCGAAAAATACCGCTGC3’ | 269 | (18) |
| 5’TCATACTCTTCCGAGCAAAGG3’ |
For detection of the aac (6’)-Ie/aph (2”), aph (3’)-IIIa, and ant (4’) genes, which are the most common aminoglycoside-modifying enzymes among the staphylococci, the PCR method was performed with specific primers (Table 1) (18). Staphylococcus aureus ATCC43300 was used as a positive control. All PCR mixtures were prepared in a 25 μL volume containing 12.5 μL of PCR master mix (Amplicon, Denmark), 1 mmol of each primer, 3 μL of DNA template, and 7.5 μL of deionized water. The mixtures were placed in a thermocycler (Pio Intellectica, Canada), and the PCR products were then visualized by electrophoresis in agarose gel, stained with ethidium bromide, and examined under UV illumination. Thermocycler programming is summarized in Table 2.
| Factor | Temperature, °C | |||
|---|---|---|---|---|
| Gene | mecA | ant(4’)-Ia | aac(6’)-Ie/aph(2”) | aph(3’)-IIIa |
| Primary denaturation | 94 | 94 | 94 | 94 |
| Denaturation | 94 | 94 | 94 | 94 |
| Annealing | 57 | 47 | 45 | 51 |
| Extension | 72 | 72 | 72 | 72 |
| Final extension | 72 | 72 | 72 | 72 |
3.5. Data Analysis
All data were analyzed with SPSS version 22. For compression of the presence of drug-resistance patterns of MRSA isolates in each area, the chi-square test was used. A P value of < 0.05 was accepted as statistically significant.
4. Results
In this study on hospitalized patients conducted during 2014, 174 MRSA isolates were obtained from various clinical samples, including blood (3.1%), sputum (6.2%), tracheal exudates (31.9%), lung (15.5%), pleura (7.1%), catheters (8%), urine (10%), and wounds (18.2%). The isolates from each hospital are described in Table 3. Drug-susceptibility testing results showed that the highest resistance in Tehran was to erythromycin (84.4%), while the highest resistance in Sari was against gentamicin (75%) (Table 4). All strains were susceptible to vancomycin. Resistance to gentamicin as an indicator of aminoglycoside resistance was observed in 71.7% of the isolates from Tehran.
| Hospital/Location (Prevalence) | Specimen Type (Prevalence) | |||||||
|---|---|---|---|---|---|---|---|---|
| Catheter (8%) | Wound (18.2%) | Urine (10%) | Pleura (7.1%) | Bronchus (15.5%) | Trachea (31.9%) | Sputum (6.2%) | Blood (3.1%) | |
| Loghman/Tehran (50.5%) | 4 | 17 | 10 | 6 | 11 | 31 | 6 | 3 |
| Pars/Tehran (28.8%) | 2 | 10 | 6 | 4 | 9 | 14 | 3 | 2 |
| Zare/Sari (20.7%) | 3 | 5 | 5 | 3 | 7 | 11 | 2 | - |
| Total | 9 | 32 | 21 | 13 | 27 | 56 | 11 | 5 |
| City | Doxycycline | Rifampin | Cefazolin | Co-trimoxazole | Gentamicin | Ciprofloxacin | Clindamycin | Erythromycin | Vancomycin |
|---|---|---|---|---|---|---|---|---|---|
| Tehran, 138 (79.3) | 71 (51.4) | 78 (56.5) | 81 (58.7) | 35 (25.4) | 99 (71.7) | 104 (75.4) | 93 (67.4) | 117 (84.8) | 0 |
| Sari, 36 (20.7) | 15 (41.7) | 14 (38.9) | 22 (61.1) | 24 (66.7) | 27 (75) | 18 (50) | 18 (50) | 23 (63.9) | 0 |
| P Value | 0.351 | 0.064 | 0.851 | 0.000 | 0.835 | 0.004 | 0.079 | 0.009 | 1 |
aValues are expressed as No. (%).
According to our results, many of the isolates were multi-drug resistant (MDR). The mecA gene was detected in all isolates (Figure 1). The aac (6’)-Ie/aph (2”) gene was detected in 134 (77%) of the 174 isolates (Figure 2). In total, the aph (3’)-IIIa and ant(4’)-Ia genes were detected in 119 (68.4%) and 122 (70.1%) of the isolates, respectively (Table 5, Figures 3 and 4).
| AME | Tehran (N = 138) | Sari (N = 36) | Total | P Value |
|---|---|---|---|---|
| aac(6΄)-Ie/aph(2˝) | 108 (78.3) | 26 (72.2) | 134 (77) | 0.505 |
| aph(3’)-IIIa | 96 (69.6) | 23 (63.9) | 119 (68.4) | 0.549 |
| ant(4’) | 104 (75.4) | 16 (44.4) | 120 (69) | 0.001 |
aValues are expressed as No. (%).
5. Discussion
In the present study, the prevalence of aminoglycoside resistance in MRSA strains in Iran was evaluated, and high resistance rates were observed. In addition, most of the isolates had MDR features. According to our results, vancomycin is still a good choice against MRSA in Iranian clinics. It is important to be aware that aminoglycoside-resistant MRSA is distributed in hospitals in Iran, and should not be neglected.
During the past few decades, S. aureus has become the most common cause of nosocomial infections (20, 21). One of the success factors of this bacteria is its resistance to drugs. With the arrival of each new antibiotic, resistant strains of bacteria have rapidly emerged and the treatment of infections caused by these strains has become difficult. For example, after the introduction of penicillin to the market for the treatment of hospitalized patients, penicillin-resistant strains quickly appeared (22). Therefore, the rapid and accurate identification of MRSA is necessary in order to help physicians make appropriate selections for antibiotic treatment, and to prevent the spread of these species (16).
According to Askari et al.’s meta-analysis and systematic review in 2012, the overall prevalence of MRSA in Iran varied from 20% to 90%, with the average prevalence in Tehran found to be 52% (23). Thus, considering the importance of the pathogenesis and increasing prevalence of MRSA in recent years, regular monitoring of the antibiotic susceptibility of isolates is necessary to control and prevent the spread of resistant strains.
Our study showed that the rate of methicillin resistance in the two investigated Tehran hospitals reached 43.5%, compared to 11.3% in the Sari hospital. Factors that may affect the prevalence of MRSA in different parts of the world include different infection-control policies, the amount of antibiotics and how they are prescribed, the predominant population or types of strains, and the laboratory methodologies used for MRSA detection (24, 25).
Despite their nephrotoxicity, ear toxicity, and problems associated with bacterial resistance, aminoglycosides continue to be effective, particularly for the treatment of staphylococcal infections (25). Cell wall synthesis inhibitors such as beta-lactams lead to increased uptake of aminoglycosides, and antibiotic combinations have useful and valuable antimicrobial properties (26). Several studies have shown a relationship between resistance to aminoglycosides and resistance to methicillin (27). In some countries, aminoglycoside resistance among S. aureus strains is common. Moreover, resistance to gentamicin is especially important because it is often used in the treatment of staphylococcal endocarditis in combination with a beta-lactam or a glycopeptide (28). Sensitivity of S. aureus isolates to gentamicin is a marker for sensitivity to other aminoglycosides. In the present study, we considered not only the aac (6’)-Ie/aph (2’) gene, which encodes the AAC (6’)-APH (2’) enzyme responsible for gentamicin resistance, but also other modifying enzymes (18, 28, 29). In this study, drug-susceptibility testing showed rates of gentamicin-resistant MRSA strains of 71.7% in Tehran and 75% in Sari.
Previous studies showed that the aac (6')/aph (2') gene is the most prevalent gene encoding AME enzymes among clinical MRSA isolates in European countries (18). Choi et al. obtained similar results in South Korea in 2003 (19). Their results showed that the prevalence of the aac (6')/aph (2') gene among MRSA isolates was 65%, followed by the aph (3')-IIIa and ant (4')-Ia genes at 41% and 9%, respectively; our results were in accordance with these. In a study conducted by Ardic and colleagues in 2006, the aac (6')/aph (2") gene was found in 66% of MRSA isolates, followed by the ant (4)-Ia and aph (IIIa) genes at 24% and 8%, respectively (30). In contrast, in a survey conducted by Ida et al. in 2001 on 381 clinical isolates in Japan, the prevalence of ant (4)-Ia was 84.5%, which was higher than the rate of two other genes (31). Yadegar et al. (2009) reported a prevalence of the ant (4')-Ia gene as 58%, higher than other modifying enzyme genes among MRSA strains isolated from Tehran hospitals (32). The present study’s results were similar to the majority of studies, in which aac (6')/aph (2'')-Ia was the predominant gene. In this case, 78.3% and 72.2% of MRSA isolates carried the aac (6')/aph (2'')-Ia gene in Tehran and Sari, respectively.
In conclusion, aminoglycoside-resistant genes are different in various MRSA colonies in different geographical regions (31). Also, the aac (6')/aph (2') gene products cause higher resistance to aminoglycosides among MRSA isolates in Iran. Moreover, we can conclude that in different geographical regions of Iran, different genes may be the cause of aminoglycoside resistance. With regard to the high rate of aminoglycoside-resistant MRSA isolates in our study, we recommend that the use of aminoglycosides against MRSA infections must be limited in Iranian hospitals. Even natural products (33) can be used to help in the treatment and eradication of such drug-resistant cases.