1. Background
Influenza pandemics occur when new influenza A subtypes get widespread among the population. Over the past few years, the global spread of highly pathogenic avian influenza A (H5N1) and the beginning of the pandemic influenza A (pH1N1) have raised concerns about the global ranking of influenza. The use of antiviral drugs (mainly oseltamivir) combined with vaccines is the response to influenza pandemics (1). Influenza A (H1N1) virus infects young and healthy adults suggesting that this group are more susceptible to the disease. It is possible that some level of cross protection antibodies exist in older people. High risk groups for this disease include pregnant women, old people, and people with serious medical illness (2).
Symptoms of pandemic influenza consist of fever, cough, body pain, headache, sore throat, and gastrointestinal complications. This illness is mostly self-limited, thus, a large proportion of infected patients do not get registered in health centers and the Statistics estimating the disease are not correct (3). The definition of influenza A (H1N1) was having high grade fever (upper than 38°C) or at least having two symptoms of respiratory disease. A confirmed case of influenza A (H1N1) was defined as a patient with high grade fever (> 38°C) or at least two of respiratory symptoms. The patient have H1N1 viral infection should be also confirmed by reverse transcriptase PCR (RT-PCR) (4-6).
Some cases have a high incidence of local transmission (such as in New York), but many other cases have led to low transmission or no secondary transmission. The mortality of influenza virus is low (2 % of hospitalized patients), but its morbidity is high (7). To control the influenza A H1N1 infection, early diagnosis with a cost-effective, rapid, and effectual assay is necessary, exclusively in developing countries. Using real time RT-PCR assay compared with other molecular techniques is recommended for the early diagnosis and the rapid identification of individuals infected with pandemic influenza virus. Fast and accurate detection of influenza improves medical handling by proper conditions of prophylaxis, rapid treatment, and appropriate management plans for public health responses to the outbreaks and the avoidance of unnecessary treatment (8).
Influenza virus infection is not detectable only with symptoms. The clinical picture of the disease Is similar to respiratory syncytial virus, parainfluenza viruses, adenoviruses, coronaviruses, and metapneumovirus (9). Correct diagnosis is urgent for the identification of pandemic influenza, surveillance, and public health interventions. Without practical laboratory tests, it is not possible to provide appropriate response efforts (10).
2. Objectives
The aim of the present study was the investigation of clinical and epidemiological figure of influenza virus A/H1N1, A/H3N2, and influenza B infection among patients with respiratory syndrome in Golestan province, the Southeast of Caspian see, Iran, 2011 to 2014.
3. Methods
3.1. Specimens
This prospective, cross sectional study took place since November 2010 through March 2014. The study population was inclusive of all suspected specimens of pandemic influenza A (H1N1) virus infection who had attended hospitals and healthcare centers in Golestan province over the study period. All patients with symptoms like influenza were included in this study and often had high grade fever (> 38°C) or any of respiratory symptoms.
Suspected cases of influenza were confirmed using a Real-time reverse-transcriptase-polymerase-chain-reaction (rRT-PCR) assay at the laboratory of influenza research center in Golestan University of Medical Sciences according to the recommended protocol of the U.S. center for disease control and prevention (CDC) (11).
In this study, the center of influenza received 790 specimens. Acceptable specimen types were given nasopharyngeal (NP) swab. Following the specimen collection in the health care center, Specimens were placed in viral transport medium and were transported to the Influenza center on cold packs.
3.2. Sample Processing and RNA Extraction
Specimens were processed for RNA extraction. The extractions were performed following the protocols supplied with high pure viral RNA extraction kit (Roche Diagnostics, Germany).
3.3. One Step Real-Time PCR Testing
After RNA extraction with High pure viral RNA extraction kit (Roche, Germany), specimens were tested for influenza A and B viruses using TaqMan probes real-time RT-PCR (rRT-PCR) according to the manual of Centers for Disease Control and Prevention (CDC) and by Invitrogen SuperScript III Platinum one step qRT- PCR kit. The subtype of influenza virus was determined for positive influenza A specimen. The subtyping consisted of A/H3 influenza (hemagglutinin gene), A pandemic (H1N1) 2009 virus (hemagglutinin gene), and Influenza B virus (nucleoprotein gene).
PCR was performed in 25 µL volumes containing 12.5 µL 2xrxn Buffer, 10 µM of each primer (40 µM of Influenza A primers), 5 pmol of probe, 0.5 µL of Rox, 2.5 unit of Enzyme mix and 5 µl of RNA template. The reaction was carried out in a 7300 ABI Real time PCR and with the following settings: 50°C for 30 minutes, 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing and extension 60°C for 30 seconds (Table 1).
| Name | Sequence (5’ > 3’) | Working Concentration, µM | |
|---|---|---|---|
| Influenza A | Primer F | GACCRATCCTGTCACCTCTGAC | 40 |
| Primer R | AGGGCATTYTGGACAAAKCGTCTA | 40 | |
| Probea | TGCAGTCCTCGCTCACTGGGCACG | 10 | |
| SW Inf A | Primer F | GCACGGTCAGCACTTATYCTRAG | 10 |
| Primer R | GTGRGCTGGGTTTTCATTTGGTC | 10 | |
| Probeb | CYACTGCAAGCCCA”T” ACACACAAGCAGGCA | 10 | |
| H3N2 | Primer F | AAGCATTCCYAATGACAAACC | 10 |
| Primer R | ATTGCRCCRAATATGCCTCTAGT | 10 | |
| Probe | CAGGATCACATATGGGSCCTGTCCCAG | 10 | |
| Influenza B | Primer F | GAGACACAATTGCCTACCTGCTT | 10 |
| Primer R | TTCTTTCCCACCGAACCAAC | 10 | |
| Probe | AGAAGATGGAGAAGGCAAAGCAGAACTAGC | 10 | |
| RNAase P | Primer F | AGATTTGGACCTGCGAGCG | 10 |
| Primer R | GAGCGGCTGTCTCCACAAGT | 10 | |
| Probe | TTCTGACCTGAAGGCTCTGCGCG | 10 |
aTaqMan probes consist of FAM reporter (5’-end with the reporter molecule 6-carboxyfluorescein) and BHQ1quencher (Blackhole Quencher 1) at the 3’-end.
bQuenched internally at a modified”T” residue with BHQ1, to prevent probe extension by Taq polymerase (11).
4. Results
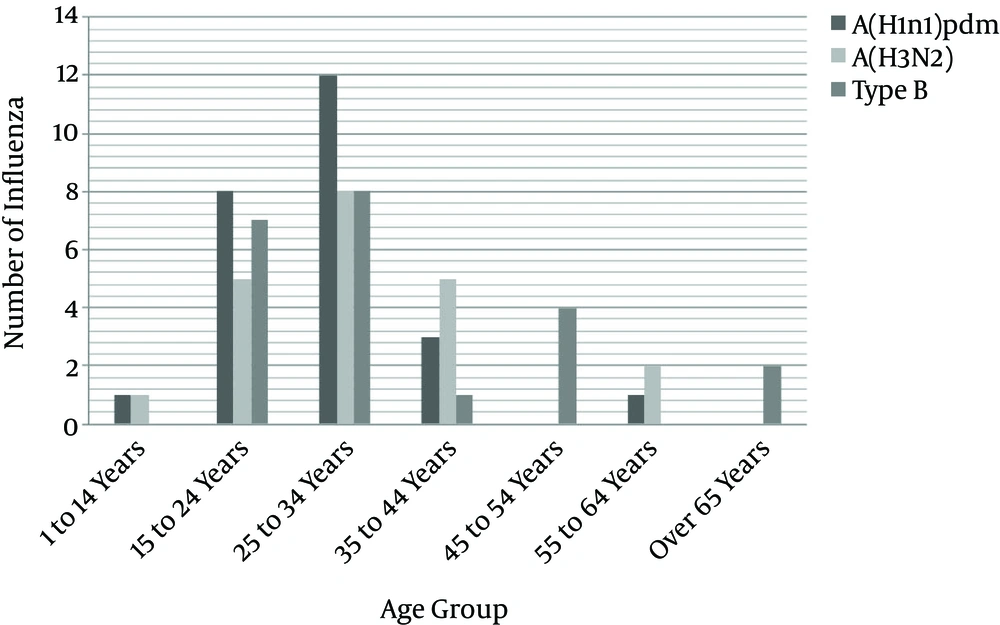
Among a total of 790 suspected cases, the following results were obtained: the number of influenza cases was 68 (8.6%) which among the proportion of influenza A/H3N2 was 21 (30.8%), influenza A/pdmH1N1 25 (36.7%), and influenza B 22(32.3%) (Table 2). Among the 790 suspected cases, male cases accounted for 72 and female cases for 718 patients. In confirmed cases 7 (10.3%) were male and 61 (89.7%) were female. The mean age was 29.79 years (SD = 16.0, ranging from < 1 days old to 92 years old). The greatest number of confirmed cases occurred in the age group of 25 to 34 years, accounting for 354 (44.8%) of all cases, followed by 227 (28.7%) in 15 to 24 years and the lowest number was in ≥ 65 years, with 22 cases (2.8%) of all cases.
| Characteristics | No. Samples Tested | No. of Influenza Cases | No. of Influenza Cases | |||
|---|---|---|---|---|---|---|
| A (H3N2) | A (H1N1) pdm09 | Type B | Type A and B | |||
| Year | ||||||
| 2010 - 2014 | 790 | 68 (8.6) | 21 (30.8) | 25 (36.7) | 22 (32.3) | 0 |
| Gender | ||||||
| Male | 72 | 7 (10.3) | 4 (57.1) | 1 (14.3) | 2 (28.6) | 0 |
| Female | 718 | 61 (89.7) | 17 (27.9) | 24 (39.3) | 20 (32.8) | 0 |
| Age group, y | ||||||
| 1 to 14 | 32 | 2 (2.9) | 1 (50) | 1 (50) | 0 | 0 |
| 15 to 24 | 227 | 20 (29.4) | 5 (25) | 8 (40) | 7 (35) | 0 |
| 25 - 34 | 354 | 28 (41.2) | 8 (28.6) | 12 (42.8) | 8 (28.6) | 0 |
| 35 to 44 | 83 | 9 (13.2) | 5 (55.6) | 3 (33.3) | 1 (11.1) | 0 |
| 45 to 54 | 42 | 4 (5.9) | 0 | 0 | 4 (100) | 0 |
| 55 to 64 | 30 | 3 (4.4) | 2 (66.7) | 1 (33.3) | 0 | 0 |
| Over 65 | 22 | 2 (2.9) | 0 | 0 | 2 (100) | 0 |
| Total | 790 | 68 (8.6) | 21 (30.8) | 25 (36.7) | 22 (32.3) | 0 |
aValues are expressed as No. (%).
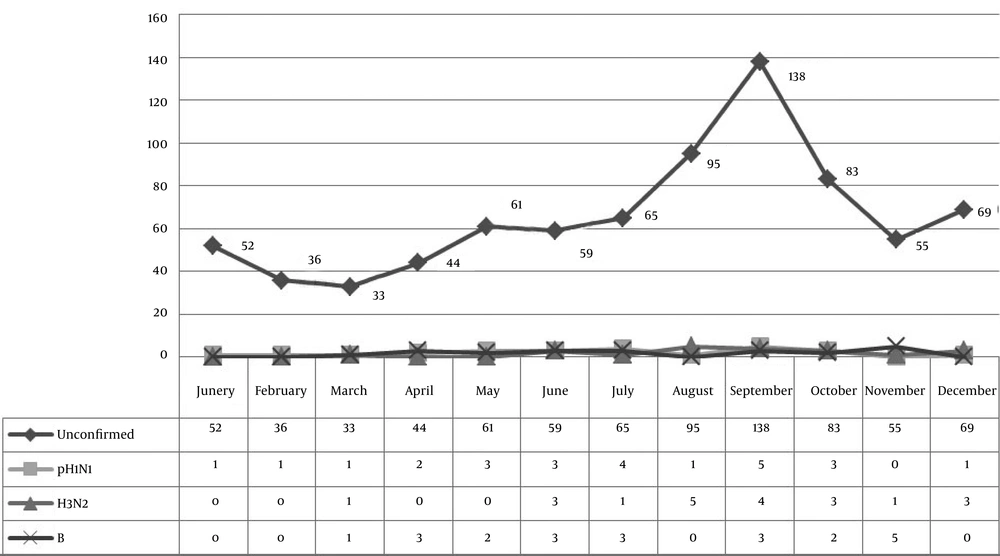
Among the total cases, 244 (30.9%) were hospitalized of which 16 (23.5%) were confirmed cases of influenza. From a total of 790 suspected cases, 283 (35.8%) were pregnant women including 28 (41.2%) confirmed cases (Tables 3 and 4). The most signs were cough (88.7%) and fever (84.3%). As shown in Figures 1 and 2, the highest incidence of confirmed cases occurred in December.
| Variable | Confirmed Cases (68 Cases) | Unconfirmed Cases (722 Cases) | P Value | |
|---|---|---|---|---|
| Sex | Male | 7 (10.3) | 65 (9) | 0.07 |
| Female | 61 (89.7) | 657 (91) | ||
| Residency | Urban | 21 (30.9) | 216 (29.9) | 0.5 |
| Rural | 47 (69.1) | 506 (70.1) | ||
| Hospitalization | Yes | 16 (23.5) | 228 (31.6) | 0.2 |
| No | 52 (76.5) | 494 (48.4) | ||
| Pregnancy | Yes | 28 (41.2) | 255 (35.3) | 0.01 |
| No | 40 (58.8) | 467 (64.7) |
aValues are expressed as No. (%).
| Positive Sample | Clinical Symptoms and Tests | ||||
|---|---|---|---|---|---|
| Fever | Cough | Myalgia | Sore throat | Rhinorrhea | |
| pH1N1 | 24 (96) | 24 (96) | 22 (88) | 10 (40) | 15 (60) |
| H3N2 | 20 (95.2) | 19 (90.4) | 16 (76.1) | 11 (52.3) | 12 (57.1) |
| B | 20 (90.9) | 21 (95.4) | 16 (72.7) | 16 (72.7) | 11 (50) |
aValues are expressed as No. (%).
5. Discussion
Influenza virus infection can spread rapidly and that is responsible for morbidity and mortality each year in the world. Mutations in the genes of the influenza virus lead to diseases with different symptoms. Therefore, it is important to identify the symptoms in different areas. Checking the symptoms can help in the diagnosis of the disease and its treatment. Chronic respiratory diseases as well as hypertension, diabetes mellitus, and pregnancy were the most common risk factors in the sever disease. Moreover, acute respiratory distress syndrome and viral pneumonia were the most common clinical causes of mortality (7, 12).
In this study, out of 790, the suspected number of influenza cases was 68 (8.6%) among which the proportion of influenza A/ pdmH1N1was 25 (36.7%). Although in Monsef’s study (2013) in Hamadan out of 180 patients, 63.8% were H1N1 positive. In Dashti-Khavidaki’s study (2009, Tehran), 34.5%, which was consistent with our results. In Afrasiabian’s study (2009, Kurdestan), 14.8%; Jedary Seifi’s study (2014 Tabriz), 20.3%; and Gao et al.’s (2009, USA), 19.5% were H1N1 positive (3, 7, 12-14). After several years of the 2009 pandemic outbreaks, H1N1pdm09 was, still, the dominant circulating strain. This was supported by world health organization in 2010 which says that the pandemic strains may circulate as seasonal viruses.
Influenza A/H3N2, in our result, was 21 (30.8%), also, in the study by Haghshenas (2011 - 2013) in which a total number of 201 patients (35.2%) were diagnosed with influenza A1 H3N2 infection. Contrary to Hajikhezri’s study which was 6.8%. Burden of influenza B in our study was 32.3%, also, in Gaini et al.’s in global influenza B study (2000 to 2013) was 22.6%, and in Qi et al.’s (2011 - 2015), influenza B was 34.1% (15-18). In the current research, fever (96%), cough (96%), and myalgia (88%) were the most common symptoms among the infected patients with pH1N1, but Rhino rhea and sore throat were present only in 60% and 40% of the patients, respectively (Table 5). These observations were in accordance with the results obtained in other studies, finally, it was found that fever, cough, and myalgia were the best diagnostic model for H1N1infection (19).
| Study | Year | Influenza Cases, % | |||
|---|---|---|---|---|---|
| A (H3N2) | A (H1N1) pdm09 | Type B | |||
| Dashti-Khavidaki et al. | 2009 | Iran-Tehran | - | 34.5 | - |
| Afrasiabian et al. | 2009 | Iran-Kurdestan | - | 14.8 | - |
| Gao et al. | 2009 | USA | - | 19.5 | - |
| Monsef et al. | 2013 | Iran-Hamedan | - | 63.8 | - |
| Jedary Seifi et al. | 2014 | Iran-Tabriz | - | 20.3 | - |
| Haghshenas et al. | 2011 - 2013 | Iran-Mazandaran | 35.2 | - | - |
| Hajikhezri et al. | 2009 - 2010 | Iran-Ahvaz | 6.8 | - | 0.7 |
| Gaini et al. | 200 - 2013 | Global study | - | - | 22.6 |
| Qi et al. | 2011 - 2015 | china | 44.7 | 21 | 34.1 |
In this study, among the confirmed cases, 7 (10.3%) were male and 61 (89.7%) were female. Also, Li et al. in 2011 in a their study reported that female subjects with influenza A are higher than males (20). Also, this point was reported by other researchers (21-23). And in Cao et al.’s study, male subjects were higher, however, Caini et al. presented that sexes were equally distributed (15). And in Cao et al.’s study, there was no sex difference in the incidence of confirmed pandemic influenza A (H1N1) (24). The mean age of the confirmed cases in the present study was 29.7 years and this was similar to other studies. In this study, there were 22 people aged > 65 years. The lower level of infection and susceptibility to influenza virus in this age group compared with younger age groups can be explained by pre-existing immunity, lower chances of being in crowded places, and fewer contacts with other people (2).
In this study, the peak incidence of the disease occurred in December and, considering the age group of samples, these peaks can be attributed to the transmission of the disease in universities and schools (2). In the last of the pandemic influenza, fast and accurate patient identification remains crucial in preventing the extensive transmission (2).
In conclusion, the outbreak of pandemic Influenza viruses in recent years has created a lot of deaths and financial losses around the world. This result shows the importance of the rapid identification of common serotypes in our society. The detection of antigenic types of circulating viruses has a great importance in vaccine preparation for high-risk individuals. Therefore, using RT-PCR is recommended for early diagnosis and rapid identification of individuals infected with pandemic influenza virus. In addition, it can accelerate vaccine manufacture. Molecular methods should be used in vaccine manufacturing in cases of acute infections. Moreover, Scheduling the urgent influenza A vaccination program is suggested, which can definitely prevent the spread of the virus.