1. Background
A leading cause of nosocomial infection, Staphylococcus aureus is responsible for many conditions, including wound infections, food poisoning, osteomyelitis, and endocarditis, as well as life-threatening diseases, such as pneumonia and bacteremia (1). This bacterium is characterized by its remarkable ability to acquire resistance to antimicrobial agents, especially methicillin. In particular, methicillin-resistant S. aureus (MRSA) has recently emerged as a major public health concern. Methicillin was the first therapeutic option developed to treat infections caused by penicillin-resistant S. aureus (2).
The first MRSA isolate was reported in 1961 in the United Kingdom (3, 4). Since then, studies have revealed a steady increase in the incidence of MRSA infection. Methicillin resistance reportedly arises from the expression of a methicillin-hydrolyzing β-lactamase or the expression of an altered form of penicillin-binding protein-2 (PBP2a, also referred to as PBP2′) that is mediated by the mecA gene. This gene is carried within a mobile genetic element known as staphylococcal cassette chromosome mec (SCCmec) (5).
MRSA infection is currently an important cause of morbidity and mortality in both community and healthcare settings due to its resistance to nearly all currently available beta-lactam antibiotics and other therapeutic options, such as macrolides, lincosides, and aminoglycoside (6, 7). The dissemination of MRSA with multi-resistance genes has significantly limited the choice of therapeutic options available to treat staphylococcal infections, which are associated with poor clinical outcomes (1, 7). Hospital-associated MRSA (HA-MRSA) strains are usually resistant to many antibiotics and may carry virulence genes, such as the pvl gene, which encodes panton-valentine leukocidin (pvl). pvl is a putative virulence factor that has been hypothesized to enhance the bacterium’s ability to cause severe infections in human and animal hosts (8).
Epidemiological studies using molecular typing are an essential component in the study of clonal relatedness, evolutionary pathways, the genetic diversity of the pathogen, and tracking the spread of S. aureus infections (9, 10). Various molecular typing methods can be used for typing MRSA isolates (10). Although pulse field gel electrophoresis (PFGE) with a high discriminatory ability is the documented gold standard among the various DNA sequence-based methods, spa typing could also be an effective and rapid method for typing MRSA isolates. spa typing is a rapid, affordable, and easy technique that offers better discriminatory abilities and is cheaper than multilocus sequence typing (MLST), which has enabled it to become a widely distributed typing technique for S. aureus isolates (11-14).
This method is based on the number of tandem repeats and the sequence variation in region X of the protein A gene. The spa gene contains three distinct regions: Fc, X, and C (12). Based on a literature review, the spa type distribution of MRSA strains isolated from patients in different geographic locations in the world exhibits a different pattern (11).
2. Objectives
The present study determined the patterns of antibiotic resistance by antibiotic sensitivity testing using different spa types of nosocomial S. aureus collected from clinical sources in Tehran, the capital city of Iran.
3. Materials and Methods
3.1. Study Design and Population
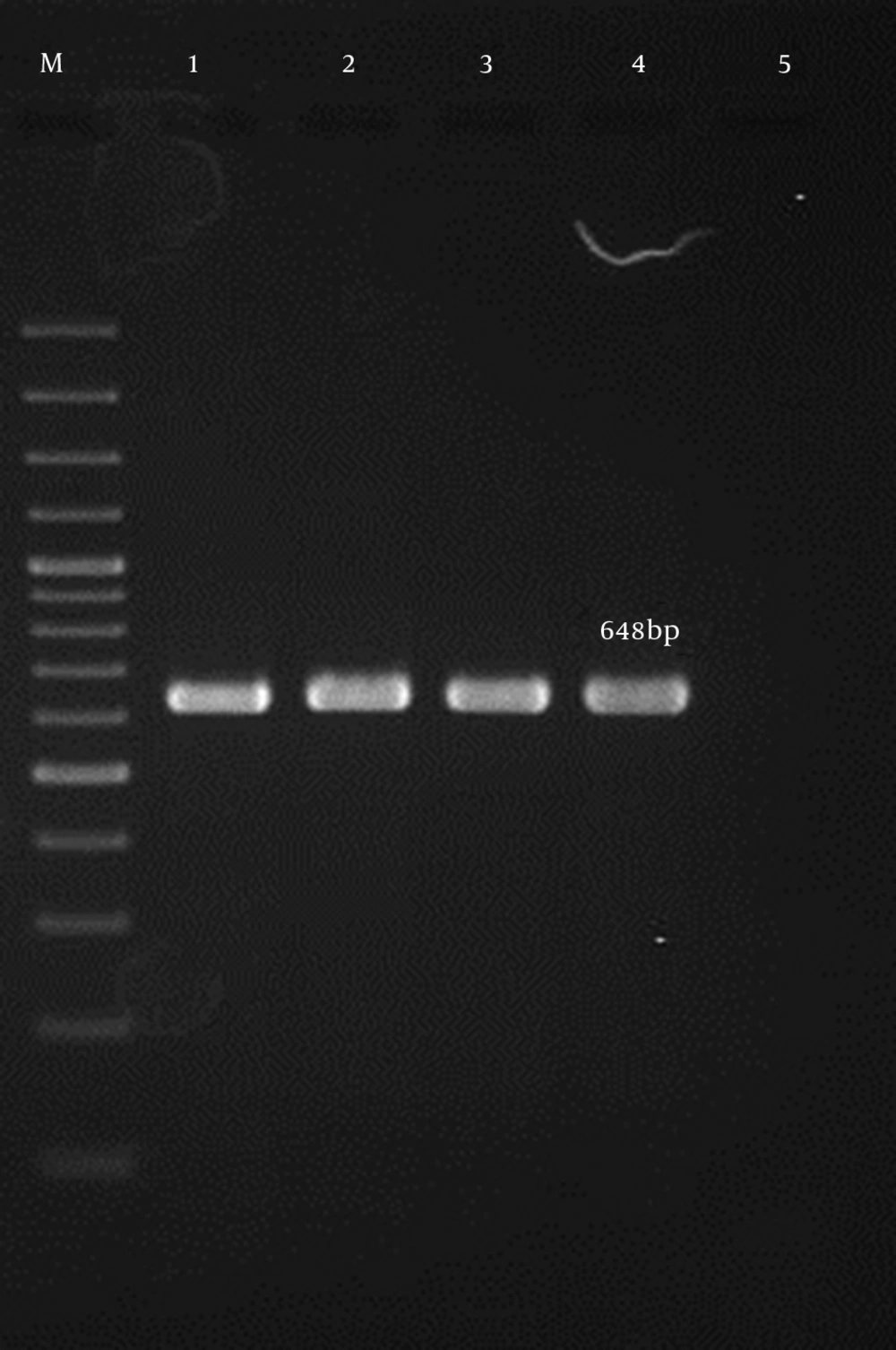
This cross-sectional study was conducted during a six-month period from the first of April 2015 to the end of September 2015. The research was approved by the ethics committee of Shahid Beheshti University of Medical Sciences [No. 1394.157]. All hospitalized patients with S. aureus infections were examined, and 90 S. aureus isolates were recovered from any clinical site of these patients. One isolate per patient was included in the study, and duplicate samples were excluded. All clinical samples were immediately transported to the laboratory upon collection. Standard microbiological procedures, such as colony morphology, Gram staining, growth on mannitol salt agar, and the production of catalase, coagulase, and DNase, were carried out for the presumptive isolation and identification of S. aureus. All isolates were confirmed using polymerase chain reaction (PCR) for the femA and nucA genes (15, 16).
3.2. Antimicrobial Susceptibility Testing
The confirmed S. aureus strains were tested for their in vitro antimicrobial resistance pattern to a panel of 16 antibiotic discs with the Kirby-Bauer disk diffusion technique on Mueller-Hinton agar (Mast, UK). The interpretive criteria for susceptibility were used by the clinical and laboratory standards institute (CLSI) (17), and the results were recorded after incubation for 18 hours at 37°C. The antimicrobial drugs tested included penicillin (PG 10 µg), ampicillin (AP 10 µg), vancomycin (VA 30 µg), teicoplanin (TEC 30 µg), ceftriaxone (CRO 30 µg), gentamicin (GM 10 µg), kanamycin (K 30 µg), amikacin (AK 30 µg), tobramycin (TN 10 µg), linezolid (LZD 30 µg), erythromycin (E 15 µg), gatifloxacin (GAT 5 µg), clindamycin (CD 2 µg), levofloxacin (LEV 5 µg), ciprofloxacin (CIP 5 µg), and trimethoprim-sulfamethoxazole (TS 25 µg). Intermediate sensitivity was scored as resistance. Multidrug resistance (MDR) was defined as resistance to three or more unique antibiotic classes in addition to beta-lactams. All antibiotic disks were obtained from Mast, UK. S. aureus ATCC25923 was used as a quality control strain in every test run. All strains were stored in Tryptic Soy Broth (TSB; Merck, Germany) that contained 20% glycerol at -80°C until use.
3.3. MRSA Screening
MRSA isolates were screened with cefoxitin (30 µg) and oxacillin discs (1 µg) on Mueller-Hinton agar plates in accordance with the CLSI guidelines (17). All methicillin-resistant isolates detected phenotypically were confirmed by PCR for the amplification of the mecA gene (18).
3.4. Genomic DNA Extraction
The QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) was used for genomic DNA extraction according to the manufacturer’s instructions. Lysostaphin (Sigma-Aldrich, US) was used to a final concentration of 15 µg/mL for cell wall lysis. The concentration of the DNA was assessed by spectrophotometry.
3.5. Detection of the Toxin-Encoding Genes
All isolates were tested for the presence of lukS-PV-lukF-PV (pvl genes) and toxic shock syndrome toxin (tsst) gene. The degenerate primers are listed in Table 1.
| Primer | Primer Sequence (5´ → 3´) | Product Size, bp | Reference |
|---|---|---|---|
| femA | 648 | (15) | |
| F | CTTACTTACTGCTGTACCTG | ||
| R | ATCTCGCTTGTTGTGTGC | ||
| nucA | 270 | (16) | |
| F | GCGATTGATGGTGATACGGTT | ||
| R | AGCCAAGCCTTGACGAACTAAAGC | ||
| mecA | 583 | (18) | |
| F | AGAAGATGGTATGTGGAAGTTAG | ||
| R | ATGTATGTGCGATTGTATTGC | ||
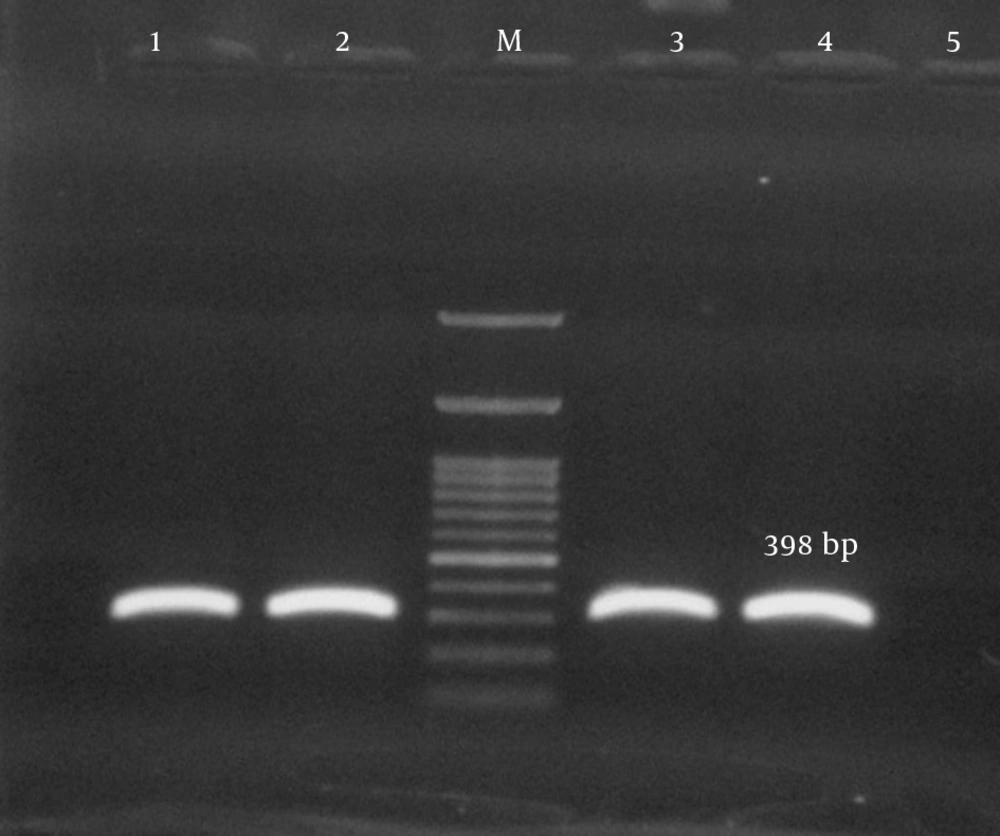
| tsst-1 | 398 | (17) | |
| F | TTATCGTAAGCCCTTTGTTG | ||
| R | TAAAGGTAGTTCTATTGGAGTAGG | ||
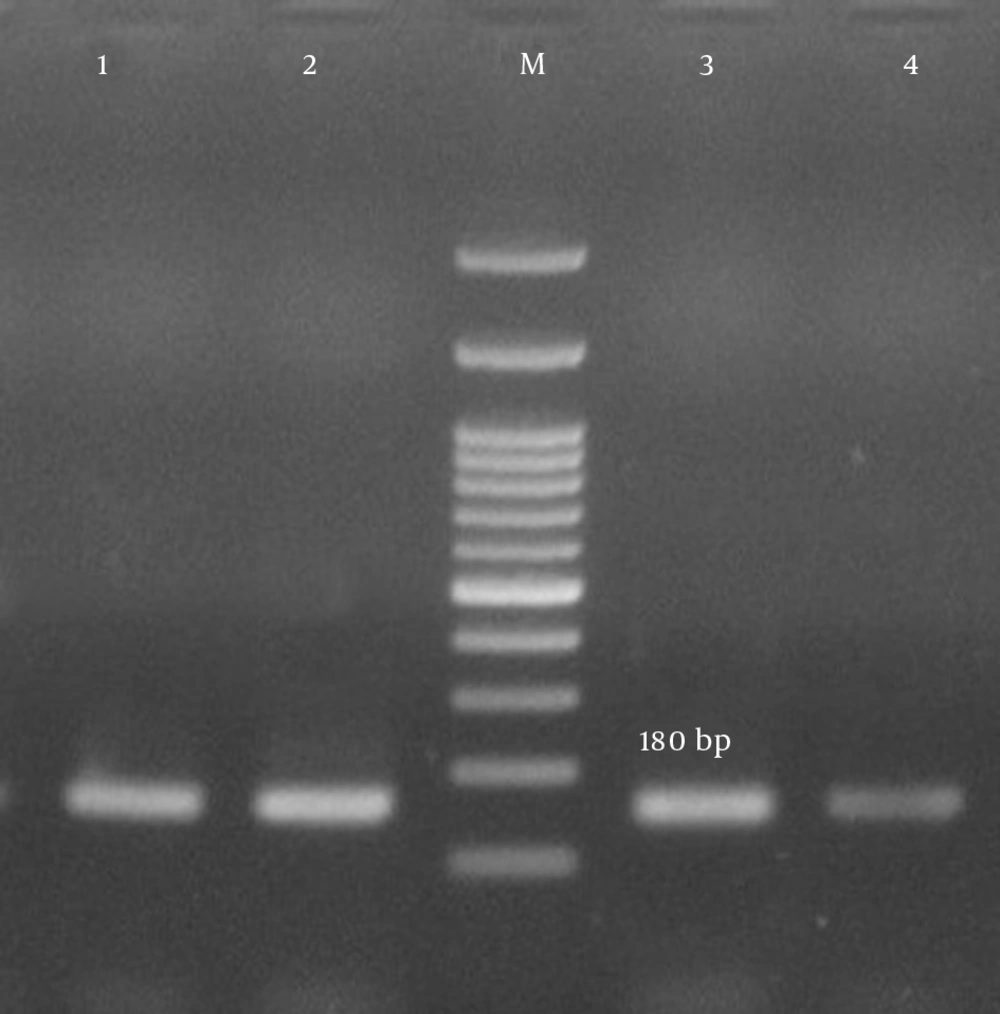
| luk-PV | 180 | (19) | |
| F | TTCACTATTTGTAAAAGTGTCAGACCCACT | ||
| R | TACTAATGAATTTTTTTATCGTAAGCCCTT |
3.6. Spa Typing
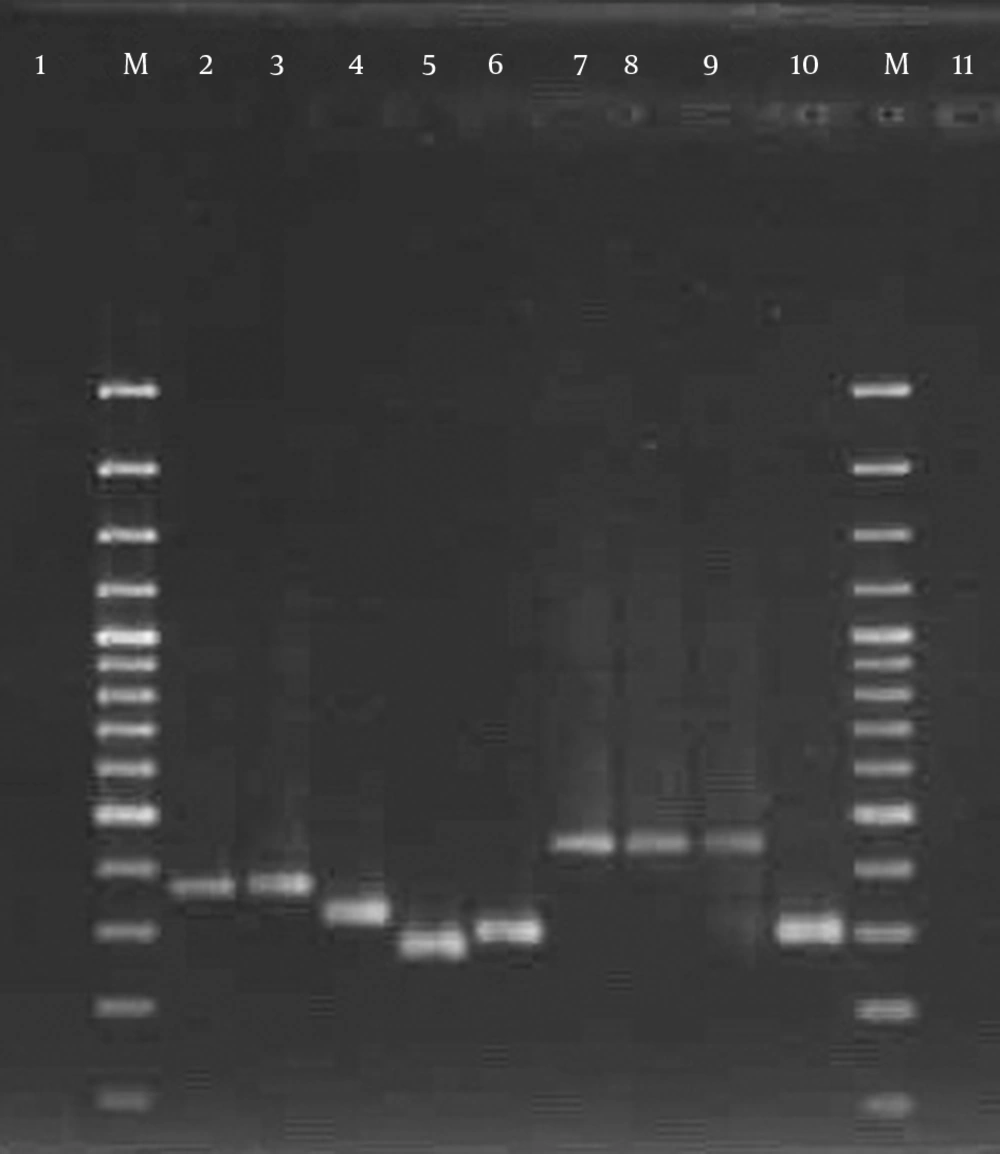
Spa typing was performed as described by Harmsen et al. (12). spa gene PCR products were subjected to DNA sequencing for both strands by Macrogen (Seoul, South Korea). The sequences obtained were edited using Chromas software (version 1.45, Australia). The guidelines from the Ridom Spa Server database (http://www.spaserver.ridom.de) were used to assign the edited sequences to particular spa types.
3.7. Statistical Analysis
Statistical analysis was performed using the statistical package for the social sciences (SPSS) for windows, version 18.0 (SPSS Inc., Chicago, IL, US).
4. Results
In all, 90 non-duplicate S. aureus isolates were obtained from 320 clinical specimens collected from hospitalized patients during a six-month period of study. All isolates were positive for the femA and nucA genes (15, 16) (Figures 1 and 2). The mean age of patients was 42 years (median: 44.1 years, range: 9 months to 71 years). The incidence of nosocomial infection with S. aureus was highest in patients from 21 - 45 years (60%) and lowest in the age group from 9 months to 20 years (3.1%).
Of the 90 analyzed S. aureus isolates, 36 (40%) were obtained from a wound, 22 (24.4%) came from the blood, 9 (10%) were collected from the ear, 9 (10%) from pus, 6 (6.7%) from body fluids, 5 (5.6%) from a catheter and, finally, three isolates (3.3%) from urine samples. The vast majority of patients was female (77.8%), while only 22.2% were male. The overall prevalence of MRSA in our study was 88.9%. The following resistance patterns were observed among our isolates: penicillin (100%), ampicillin (100%), erythromycin (82.2%), ciprofloxacin (76.7%), amikacin (65.6%), gentamicin (63.3%), clindamycin (60%), kanamycin (55.6%), tobramycin (50%), gatifloxacin (50%), trimethoprim-sulfamethoxazole (44.5%), levofloxacin (32.2%), ceftriaxone (23.3%), vancomycin (0%), teicoplanin (0%), and linezolid (0%). The distribution of the different clinical samples and their resistance profiles in MRSA isolated from patients are summarized in Table 2.
| Type of Clinical Infections | Resistance to Antibiotics | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PG | AP | CRO | GM | K | AK | TN | E | GAT | CD | LEV | CIP | TS | ||
| Wound | 36 (100) | 36 (100) | 12 (33.3) | 28 (77.8) | 30 (83.3) | 28 (77.8) | 10 (27.8) | 36 (100) | 26 (72.2) | 33 (91.7) | 10 (27.8) | 35 (97.2) | 20 (55.6) | 36 (40) |
| Blood | 22 (100) | 22 (100) | 8 (36.4) | 15 (68.2) | 12 (54.5) | 20 (90.9) | 18 (81.8) | 20 (90.9) | 11 (50) | 19 (86.4) | 6 (27.3) | 20 (90.9) | 16 (72.7) | 22 (24.4) |
| Ear | 9 (100) | 9 (100) | 0 (0) | 2 (22.2) | 0 (0) | 2 (22.2) | 5 (55.6) | 4 (44.4) | 6 (66.7) | 0 (0) | 4 (44.4) | 5 (55.6) | 0 (0) | 9 (10) |
| Pus | 9 (100) | 9 (100) | 1 (11.1) | 1 (11.1) | 0 (0) | 9 (100) | 4 (44.4) | 3 (33.3) | 0 (0) | 2 (22.2) | 3 (33.3) | 5 (55.6) | 2 (22.2) | 9 (10) |
| Body fluids | 6 (100) | 6 (100) | 0 (0) | 4 (66.7) | 5 (83.3) | 0 (0) | 2 (33.3) | 6 (100) | 0 (0) | 0 (0) | 1 (16.7) | 2 (33.3) | 1 (16.7) | 6 (6.7) |
| Catheter | 5 (100) | 5 (100) | 0 (0) | 4 (80) | 3 (60) | 0 (0) | 4 (80) | 5 (100) | 0 (0) | 0 (0) | 3 (60) | 2 (40) | 1 (20) | 5 (5.6) |
| Urine | 3 (100) | 3 (100) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 2 (66.7) | 0 (0) | 2 (66.7) | 0 (0) | 2 (66.7) | 0 (0) | 0 (0) | 3 (3.3) |
| Total | 90 (100) | 90 (100) | 21 (23.3) | 57 (63.3) | 50 (55.6) | 59 (65.6) | 45 (50) | 74 (82.2) | 45 (50) | 54 (60) | 29 (23.3) | 69 (76.7) | 40 (44.5) | |
Abbreviations: AK, amikacin; AP, ampicillin; CD, clindamycin; CIP, ciprofloxacin; CRO, ceftriaxone; E, erythromycin; LEV, levofloxacin; K, kanamycin; GAT, gatifloxacin; GM, gentamicin; PG, penicillin; SYN, quinupristin-dalfopristin; TN, tobramycin; TS, trimethoprim-sulfamethoxazole.
aValues are expressed as No. (%).
Antimicrobial susceptibility testing revealed that the rate of multidrug resistance among our isolates was 93.3%. The predominant resistance profiles in our isolates included 10 antibiotics (33.3%), 7 antibiotics (33.3%), 8 antibiotics (21.1%), and 9 antibiotics (5.6%) respectively. Twenty-three isolates (25.5%) were positive for pvl-encoding genes (Figure 3). From among the S. aureus isolates analyzed in the current study, 28 (31.1%) harbored the tsst-1 encoding gene, which was detected in wound (35.7%), blood (28.6%), pus (17.9%), catheter (10.7%), and body fluid (7.1%) samples. tsst genes were confirmed in the isolates with spa types t790 (53.6%), t044 (25%), and t037 (21.4%) (Figure 4). Fifteen isolates carried the pvl and tsst-1 genes simultaneously.
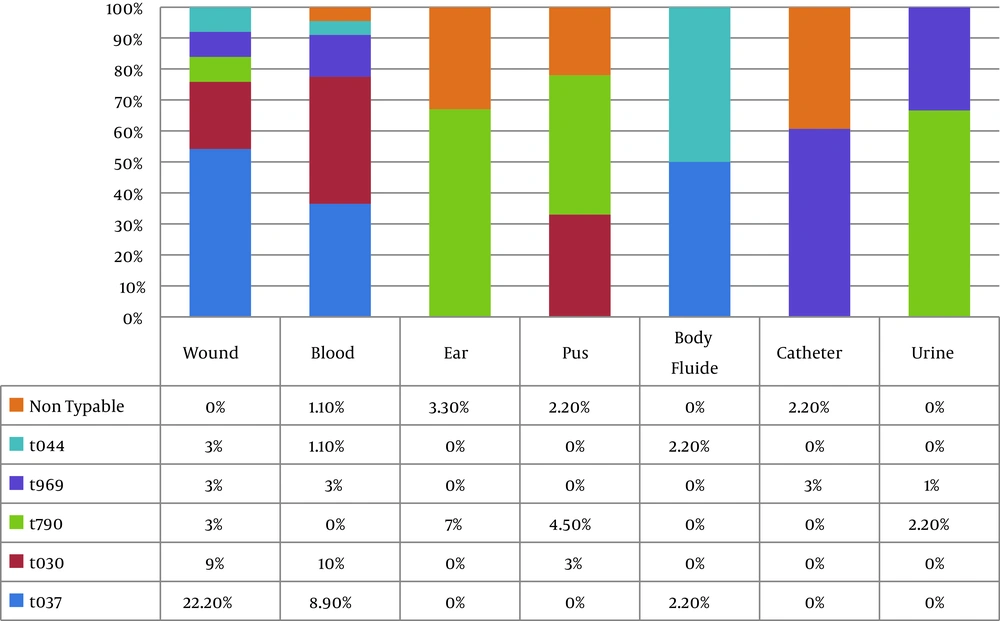
All but eight (8.9%) isolates were typable using the spa typing method. spa typing of S. aureus isolates revealed five different spa types (t037, t030, t790, t969, and t044) that were common among 30 strains (33.3%), 20 strains (22.2%), 15 strains (16.7%), 10 strains (11.1%), and 7 strains (7.7%), respectively (Figure 5). Our results indicated that all strains (100%) with spa type t044 were pvl- and tsst-positive, while the pvl-encoding gene was detected in 8 strains (80%) with spa t969, 5 strains (16.7%) with spa t037, and 3 strains (15%) with spa t030. The pvl-encoding gene was not confirmed in any of the t790 strains, while all isolates with spa t790 were positive for tsst. The spa types were obtained from different clinical samples. The distribution of spa types isolated from clinical sources is shown in Figure 6. Interestingly, 60% of the t037 isolates were found to be resistant to 10 antibiotics. All t030 isolates were obtained from patients 20 years old or younger and showed variability in their MDR patterns. Information about multiple antibiotic resistance patterns among the distribution of spa types is shown in Table 3.
| Number of Antibiotics | Resistance Pattern | No. (%) | Spa Types, No. (%) |
|---|---|---|---|
| 7 | |||
| PG, AP, E, AK, CD, CIP, GM | 20 (23.8) | t030; 5 (6), t037; 5 (6), t790; 5 (6), t969; 3 (3.6), t044; 1 (1.2), NT; 1 (1.2) | |
| PG, AP, AK, CD, TN, GAT, LEV | 6 (7.2) | t030; 2 (2.4), t969; 2 (2.4), t044; 1 (1.2), NT; 1 (1.2) | |
| PG, AP, CD, K, GAT, TS, CRO | 1 (1.2) | t030; 1 (1.2) | |
| PG, AP, AK, GM, CD, GAT, LEV | 3 (3.6) | t969; 2 (2.4), NT; 1 (1.2) | |
| 8 | PG, AP, E, CIP, CD, GM, K, TN | 19 (22.7) | t030; 3 (3.6), t037; 3 (3.6), t790; 7 (8.3), t969; 1 (1.2), t044; 5 (6) |
| 9 | PG, AP, E, CD, GAT, TS, TN, LEV, CRO | 5 (6) | t030; 1 (1.2), t037; 2 (2.4), t790; 1 (1.2), NT; 1 (1.2) |
| 10 | |||
| PG, AP, E, CIP, AK, GM, K, TN, TS, GAT | 15 | t037: 10 (12), t030; 4 (4.8), t790; 1 (1.2) | |
| PG, AP, E, CIP, AK, K, GAT, TS, LEV, CRO | 15 | t037; 8 (9.5), t030; 6 (7.1), t790; 1 (1.2) |
Abbreviations: AK, amikacin; AP, ampicillin; CD, clindamycin; CIP, ciprofloxacin; CRO, ceftriaxone; E, erythromycin; GAT, gatifloxacin; GM, gentamicin; K, kanamycin; LEV, levofloxacin; PG, penicillin; VA, vancomycin; TEC, teicoplanin; TN, tobramycin; LZD, linezolid; TS, trimethoprim-sulfamethoxazole; NT; non typable.
5. Discussion
The widespread emergence of MDR S. aureus is becoming a great public health challenge. Currently, the spread of MDR S. aureus limits therapeutic options and causes severe morbidity and mortality in hospitalized patients (20). The prevalence of MRSA also varies widely in different geographic regions of the world (21-23).
The rate of methicillin resistance in our study was 88.9%, genotypically. This result is consistent with the findings of previous studies in Iran (24) and India (25) and is higher than the rate found in Taiwan (26), Hungary (27), Serbia (28), and Croatia (29). These differences could be attributed to the studied population, the type of clinical isolates, and the trends for prescribing certain antibiotics in different geographic areas.
The results of susceptibility testing revealed that all isolates were resistant to penicillin and ampicillin yet were susceptible to vancomycin, teicoplanin, and linzolid. A similar resistance pattern was previously reported in Italy (8), Croatia (29), and Turkey (30). Based on in vitro susceptibility data, high proportions of the isolates were resistant to erythromycin (82.2%), ciprofloxacin (76.7%), amikacin (65.6%), gentamicin (63.3%), clindamycin (60%), kanamycin (55.6%), tobramycin (50%), and gatifloxacin (50%) but had a relatively low resistance to trimetoprim-sulfamethoxazole (45%), levofloxacin (32.2%), and ceftriaxone (23.3%). The results of our study support the findings of other studies (10, 31). Differences in the susceptibility pattern can be attributed to inappropriate antibiotic prescriptions, surveillance, and infection control programs in healthcare settings and also the spread of antibiotic resistance genes among bacteria. We reported a considerable increase in the prevalence of MDR (93.3%). The incidence of MDR varies widely among nations and can vary from 83.9% in Serbia (28) to 75.8% in Taiwan (26).
In this study, the spa typing method was used to observe five different spa types among our isolates: t037 (33.3%), t030 (22.2%), t790 (16.7%), t969 (11.1%), and t044 (7.7%). We found that spa type t037 was the most common spa type among our isolates. These spa types were previously described in a study conducted on S. aureus isolated from patients, personnel, the air, and the environment of an intensive care unit in Iran in 2014. In this previous study, 37 S. aureus isolates were examined for spa typing, and 11 different spa types were identified (t7688, t7689, t030, t325, t7685, t037, t297, t3096, t044, t7789). The majority of the isolates belonged to spa types t030 and t037 (43%) (24).
In a study conducted by Chen et al. to understand the molecular evolution of MRSA during a 15-year period from 1994 - 2008, the authors investigated 466 non-duplicate S. aureus isolates, including 302 MRSA and 164 methicillin-susceptible (MSSA) isolates. Chen et al. showed that from 1994 - 2000, the most predominant MRSA spa type was spa t037, while spa t030 has rapidly replaced t037 since 2000; the most obvious difference between them was resistance to rifampin (22). The resistance patterns of spa t037 in our study were in concordance with the report of Chen et al. (22), and most of the t037 strains were resistant to tetracycline, erythromycin, clindamycin, gentamicin, chloramphenicol, trimethoprim-sulfamethoxazole, and ciprofloxacin and also susceptible to rifampin and vancomycin. With a variability in its resistance pattern, t030 was the second most common spa type among our clinical isolates. This result is in concordance with the findings of some other investigators (22, 24). Chen et al. showed that t030 was the most frequent clone, accounting for 52.0% of the 302 MRSA isolates. These authors believed that t030 had a strong survival advantage and could be easily transmitted. This spa type increased significantly and has successfully become established as the dominant spa type in Chinese hospitals (22).
According to the results of the present study, t790 was the third most common spa type in Tehran. These spa types were previously described in a study conducted by Japoni-Nejad and colleagues, who analyzed the molecular characterization of CA-MRSA S. aureus strains from central Iran (9). Though several studies have reported t790 as the predominant spa type (9, 21), other observations on the limited frequency of t790 in different geographic areas also exist (22). Given the high prevalence of t790 in this study, our present findings support the view that this type could be linked to the transfer of S. aureus from the community to hospitals. In our study, we observed a low frequency of the t969 and t044 spa types along with high MDR rates in our isolates. Previous studies in other countries have also reported a low frequency of t969 and t044 spa types in comparison with other spa types, which is in accordance with our results but is not to the same extent (23, 24, 29, 32).
A major strength of the study was that it was performed on S. aureus strains isolated from the clinical specimens of patients to determine of antibiotic resistance pattern, the toxin profile, and different spa types of nosocomial S. aureus; however, the main limitation of this study was its modest sample size and the difficulty with using other methods, such as PFGE and MLST.
5.1. Conclusion
Our study reported a considerable increase in the prevalence of MDR. Based on spa typing, five distinct spa types of S. aureus were identified in our study; spa t030 and t037 were widely disseminated. Therefore, future studies should focus on identifying MDR and the prevalence of different S. aureus spa types. Infection control measures along with continuous and nationwide MRSA surveillance studies should be continued to reduce the emergence of multi-resistant strains.