1. Background
Staphylococcus aureus remains the most common pathogen, causing hospital- and community-acquired infections worldwide (1, 2). This bacterium is considered to be a colonizer in about one-third of healthy humans and is most likely found in the posterior vestibules of human nares (3). However, this colonizer could be the source of an endogenous staphylococcal infection if a host’s immune defense is compromised (4, 5). In addition, the increasing trend of resistance among S. aureus to macrolide-lincosamide-streptogramin B (MLSB) antibiotics in particular has been observed worldwide for the past few decades. For instance, persistently high prevalence rates of the inducible MLSB (iMLSB) phenotype were reported to be 94.0% in 2007 and 91.0% in 2015 in Japan (6, 7). This is not surprising because prevalence rates vary by geographical locations, by the type of strains being studied, and due to the local antibiotic policy (6, 8, 9).
The ability of this bacterium to confer cross-resistance to MLSB antibiotics is of great concern, especially for clinicians who are managing patients because this resistance may lead to clinical failure (10). For instance, isolates that exhibit resistant to erythromycin (a macrolide) but that are susceptible to clindamycin (a lincosamide) should be cautiously interpreted. This is because the presence of erm genes could mediate cross-resistance to one of the MLSB antibiotics and because this strain exhibits the iMLSB phenotype (11-13).
Strains with the iMLSB phenotype demonstrate resistance to erythromycin but are falsely susceptible to clindamycin in vitro (14). Conversely, all erythromycin-resistant isolates cannot be interpreted as being resistant to clindamycin because isolates with the MS phenotype would confer resistance to erythromycin but would be susceptible to clindamycin (15). These problems, however, can be solved by performing the double-disk diffusion (D-test) before the clindamycin susceptibility result is released to clinicians in order to detect strains that may become resistant during treatment (16). For these reasons, investigating the local prevalence rate of MLSB resistance isolates using the D-test as a phenotypic screening method and genotypically confirming the presence of its resistance genes by polymerase chain reaction (PCR) are of paramount importance. Our findings could hopefully provide new insight into the management of S. aureus infections in Malaysian hospitals.
2. Objectives
Because of the growing concern of the increasing trend of MLSB resistance among methicillin-susceptible S. aureus (MSSA) isolates in other countries, its potential clinical failure, and the limited data on this problem in Malaysia, the present study was conducted to determine the prevalence of MLSB resistance using the D-test as a phenotypic screening method and to genotypically detect the ermA, ermB, ermC, and msrA genes using PCR among MSSA isolates obtained from hospitalized patients and healthy carriers.
3. Methods
3.1. Data Collection
A total of 183 MSSA isolates from healthy carriers (50 isolates) and hospitalized patients (133 isolates) were retrieved in our laboratory during previous local studies by Ghasemzadeh-Moghaddam et al. and Neela et al. (17, 18). Duplicative isolates were excluded. The clinical isolates were obtained in 2008 from hospitalized patients in different isolation sites in various wards of the largest tertiary Malaysian hospital, which had 81 wards and 2502 beds. The carrier isolates were obtained from nasal swabs belonging to normal healthy individuals in the same year. Isolates obtained from stock cultures were stored at -80°C in a Luria-Bertani broth (Difco laboratories, USA) supplemented with 20% glycerol and were thawed accordingly during the experiment.
3.2. Identification of Bacteria
In this study, all strains were reconfirmed by standard methods, including Gram staining, catalase production, a cefoxitin test, tube coagulase testing, mannitol testing, and species-specific PCR (19). MSSA isolated from the nasal swabs of healthy individuals or carriers were defined as colonizers (20). Approval to conduct the study was obtained from the institutional ethical committee (UPM/TNCPI/RMC/1.4.18.1 JKEUPM/F1).
3.3. The D-Test
The susceptibility test against erythromycin and clindamycin was carried out using the disk diffusion method according to the guidelines of the clinical and laboratory standards institute (CLSI) (21). The erythromycin resistance isolates were further tested for the minimum inhibitory test using the E-test method according to CLSI guidelines (21). Testing for all resistance phenotypes, such as iMLSB, constitutive MLSB (cMLSB), and MS phenotypes, were accomplished using the agar disk diffusion method, in which erythromycin (15 µg) and clindamycin (2 µg) disks were placed 26 mm apart, as recommended by the CLSI, and interpreted accordingly (21). Briefly, bacterial suspensions adjusted at 0.5 McFarland were prepared and inoculated into Muller–Hinton agar (Oxoid Ltd., UK). The D-test was conducted on each plate, and finally, all plates were incubated for 18 hours at 35°C.
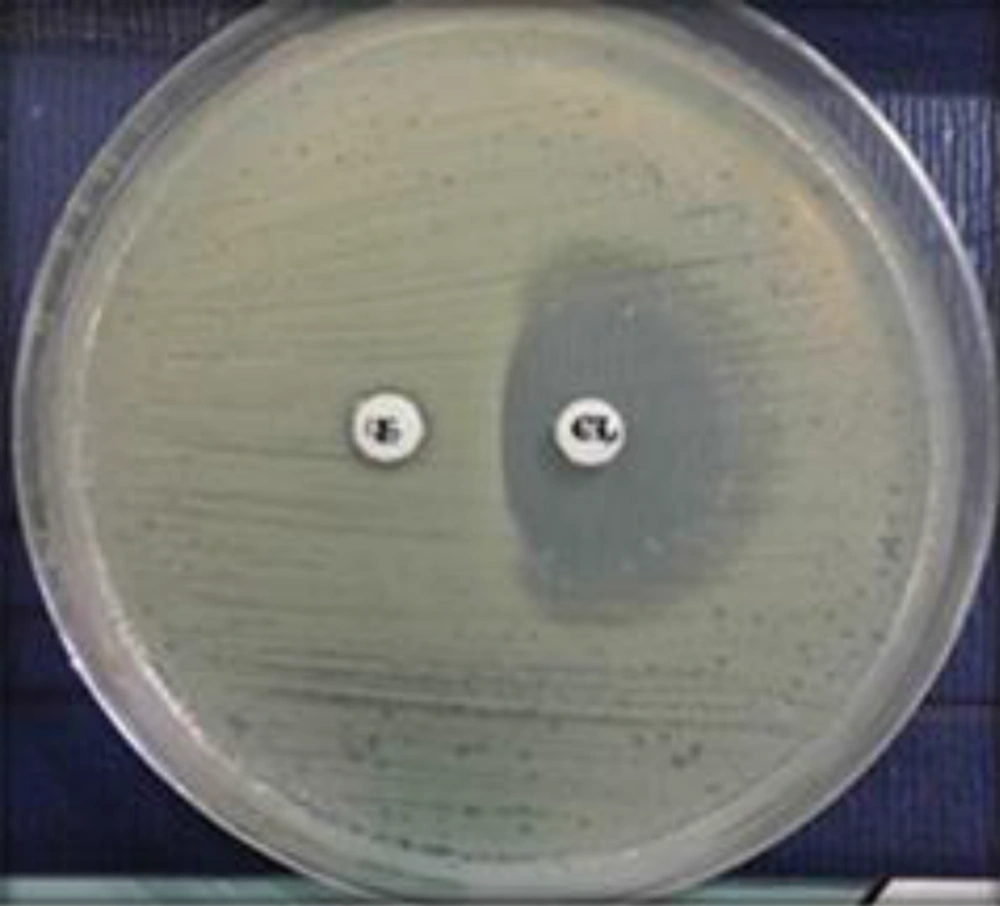
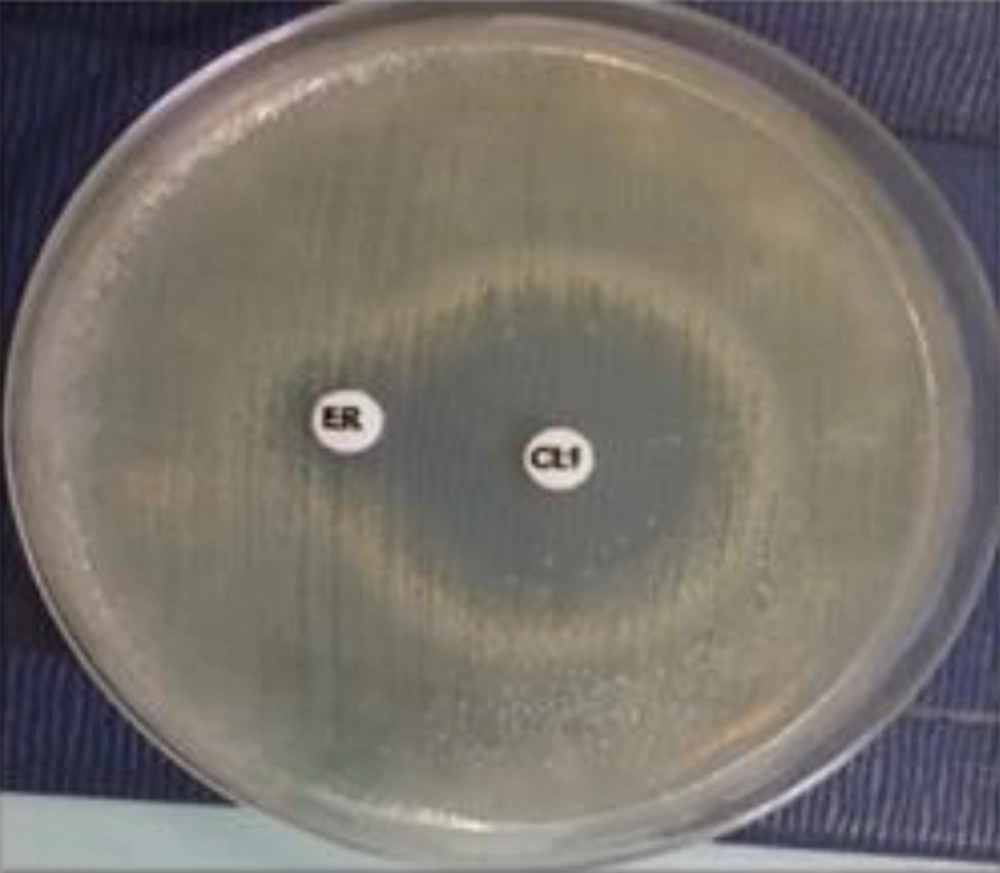
The interpretations were made according to CLSI guidelines as follows: The cMLSB phenotype was identified when there was an absence of an inhibition zone around both erythromycin and clindamycin disks, iMLSB was identified when there was a flattening or D-shaped zone around the clindamycin disk facing the erythromycin disk, and the MS phenotype was identified when an isolate was susceptible to clindamycin but was resistant to erythromycin. Staphylococcus aureus ATCC BAA 977 was used as a positive control for the D-zone.
3.4. Detection of erm and msrA Genes by PCR
Genes encoding MLSB resistance (ermA, ermB and ermC) and MS resistance (msrA) were detected by PCR using the primers described previously (11, 22, 23). The sequence of the primers and the size of PCR products are shown in Table 1.
| Primers | Sequence (5’ - 3’) | Product Size, bp | Reference |
|---|---|---|---|
| ermA | Forward: TATCTTATCGTTGAGAAGGGATT | 139 | (22) |
| Reverse: CTACACTTGGCTGATGAAA | |||
| ermB | Forward: CTATCTGATTGTTGAAGAAGCATT | 141 | (22) |
| Reverse: GTTTACTCTTGGTTTAGGATCAAA | |||
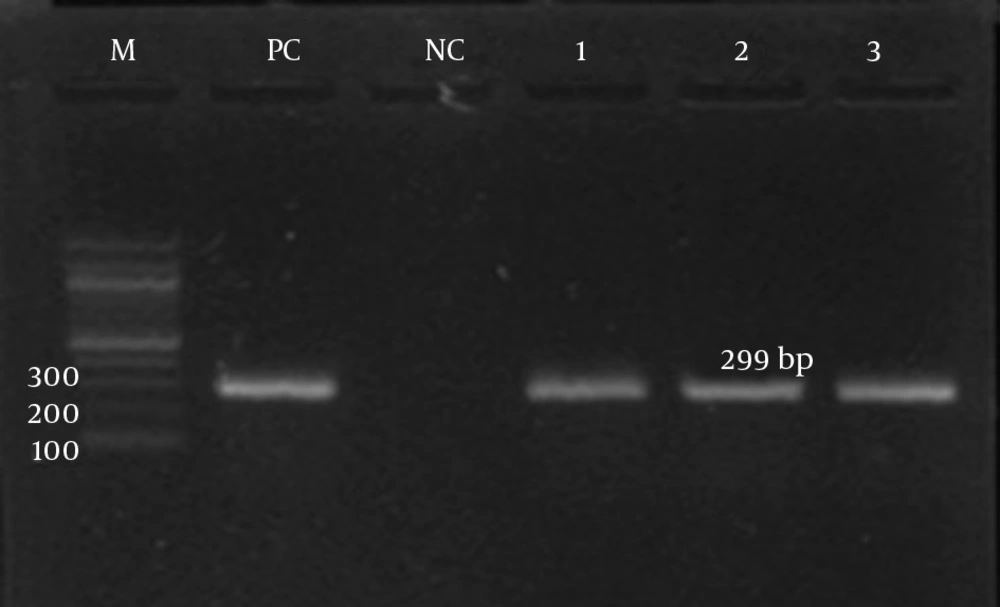
| ermC | Forward: AATCGTCAATTCCTGCATGT | 299 | (23) |
| Reverse: TAATCGTGGAATACGGGTTTG | |||
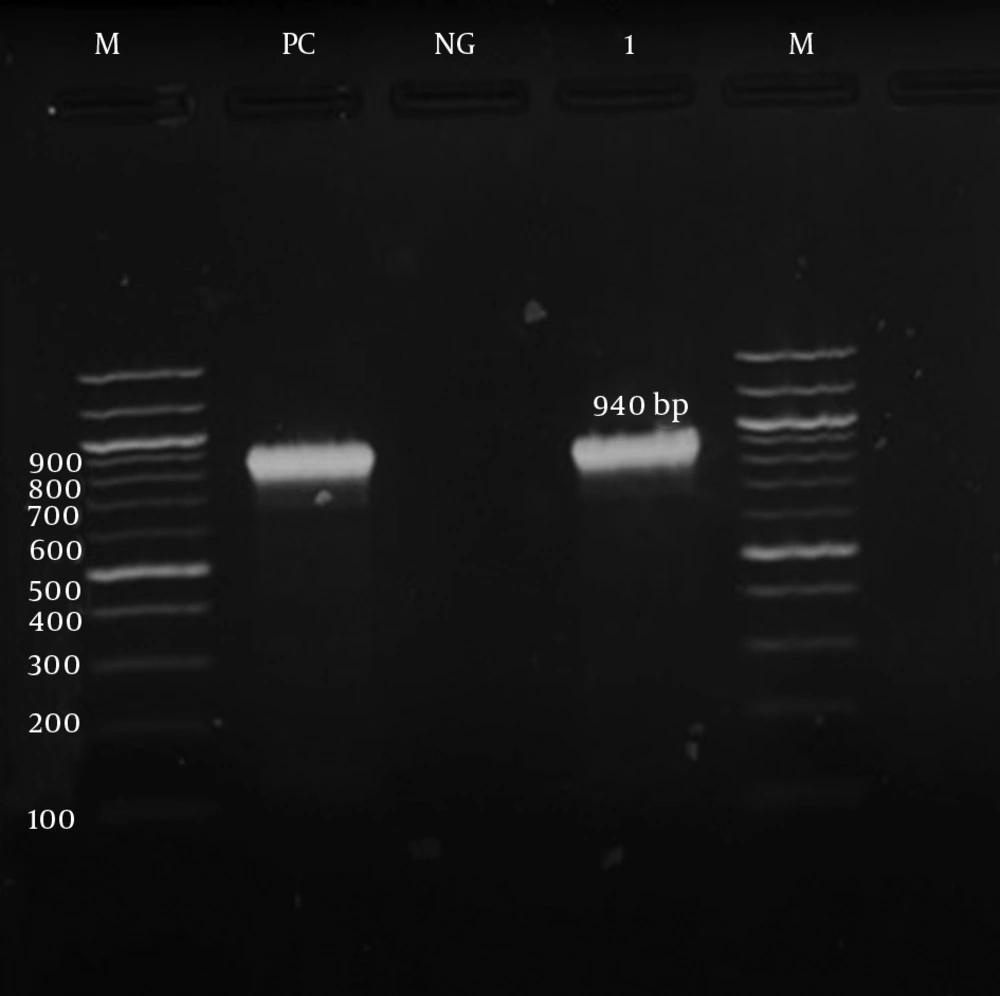
| msrA | Forward: TCCAATCATTGCACAAAATC | 940 | (24) |
| Reverse: AATTCCCTCTATTTGGTGGT | |||
| nuc | Forward: GCGATTGATGGTGATACGGTT | 279 | (25) |
| Reverse: AGCCAAGCCTTGACGAACTAAAGC |
Each individual reaction was carried out in a final volume of 25 μL that consisted of 11 μL of GOtaq green master mix (Promega, United States), 11 μL of distilled water, 1 μL of both forward and reverse primers, 1 μL of DNA, and distilled water. Thermocycling conditions included a 4-minute denaturation step at 94°C and 30 cycles of amplification with each cycle consisting of a 1-minute denaturation step at 94°C, a 1-minure annealing step at 50°C for both ermA and ermB genes, a 45-second initial extension step n at 72°C. The final extension step was extended for 7 minutes at 72°C, and 4°C was used as the holding temperature.
Streptococcus pneumoniae ATCC 700677 was used as a positive control for ermB, and Staphylococcus aureus BAA ATCC 977 was used as a positive control for ermA. Staphylococcus aureus isolates with positive results for ermC (accession no: AB982226.1) and msrA (accession no: CP010890.1) were sequenced and analyzed by BLAST and Chromas and DDBJ/EMBL/ Gen Bank. These isolates were utilized as positive controls for the identification of the respective genes. Finally, the gel was stained with Gel Red (Biotium Inc., California), and the PCR products, placed on 1.5% agarose gel electrophoresis, were photographed and analyzed using the gel documentation system (BioRad, Singapore).
4. Results
All 183 isolates were reidentified as MSSA based on their biochemical test results, their susceptibility to a cefoxitin disk, and species-specific PCR. In general, all MSSA isolates were highly susceptible to erythromycin (97.8%) and clindamycin (98.4%). Interestingly, all MSSA isolates from carriers were susceptible to both antibiotics (100%). Of the 133 MSSA isolates from hospitalized patients, 97% and 97.7% were susceptible to erythromycin and clindamycin, respectively, and only four (3%) MSSA isolates were resistant to erythromycin.
4.1. MLSB Phenotypic Resistance Patterns
Erythromycin-resistant MSSA isolates were subjected to a D-test and demonstrated MLSB resistance (2.2%). With regard to MSLB phenotypes, the D-zone phenomenon was detected in 3 (1.6%) MSSA isolates, as shown in Figure 1. Only one isolate (0.6%) demonstrated the MS phenotype, as shown in Figure 2. No cMLSB was detected in these isolates.
4.2. Prevalence of Resistance Genes
The detection of ermA, ermB, ermC, and msrA genes was investigated by PCR according to specific primers. Only the ermC gene was detected in the iMLSB phenotype strains (100%). Meanwhile, the msrA gene was detected in the MS phenotype. Both the ermC and msrA genes were detected on the gel electrophoresis, as shown in Figures 3 and 4, respectively. The ermA and ermB genes were not detected in the present study.
5. Discussion
The increasing trend of MLSB antibiotic resistance among S. aureus has gained increasing attention from scientists worldwide. The ability of strains with the iMLSB phenotype to gradually change to the cMLSB phenotype during therapy has further complicated this misery (10, 12). This is due to the fact that resistance genes, such as ermA, ermB and ermC, in iMLSB phenotype strains mediate cross-resistance to one of the MLSB antibiotics (11). These genes can also easily transferable to similar or other species through mobile genetic elements, such as transposon and plasmid (26).
There is also a challenge for clinicians with regard to treatment. Clindamycin has frequently been used for treating invasive S. aureus infections due to its good tissue penetration and other biological properties (27). However, therapeutic failures of clindamycin have been reported in patients harboring erythromycin-resistant S. aureus isolates (28). Moreover, routine in vitro tests for clindamycin susceptibility used in most diagnostic laboratories have frequently failed to recognize inducible clindamycin resistance (14). Data on the prevalence of MLSB phenotypes in Malaysia is still lacking. This prevalence may vary according to geographical locations, local institutions, type of strains, and local antibiotic policy (6, 8, 9). Thus, the present study was conducted to determine the distribution of MSLB phenotypes among MSSA isolated from hospitalized patients and carriers. The resistance genes ermA, ermB, ermC and msrA were also investigated.
In this present study, MSSA isolates were still highly susceptible to erythromycin and clindamycin. High rates of susceptibility to these antibiotics could be due to lower selective pressure among MSSA isolates. Our findings corroborate a study in Bosnia–Herzegovina in which 98% and 96.9% of MSSA isolates obtained from healthy carriers and patients, respectively, were susceptible to both antibiotics (29). In contrast, our findings were higher than those reported in other countries. Two studies from Turkey reported 89.1% and 77% susceptibility among their isolates (13, 30). In Japan, 70.5% of isolates were susceptible to both antibiotics (7). In terms of MLSB resistance, an extremely low prevalence of MLSB resistance (2.2%) was reported in the present study. Otsuka et al. reported a higher prevalence rate (34.6%) among their MSSA isolates (6). Additionally, other findings in India and Turkey reported 18.5% and 10.9% MSLB resistance, respectively, among their isolates (13, 28).
The iMLSB phenotype is more common among MSSA isolates than the cMLSB phenotype worldwide (11). In contrast, cMLSB is very common in methicillin-resistant S. aureus (6). Among the MLSB resistance isolates in the present study, 1.6% of the isolates exhibited the iMLSB phenotype. Surprisingly, data in Turkey and Japan reported higher prevalence rates of 94.4% and 94%, respectively (6, 31). However, studies in Bosnia-Herzegovina and Turkey reported lower prevalence rates of 3.5% and 8%, respectively (29, 31).
In terms of the MS phenotype, only a 0.6% prevalence rate was reported in the present study. Lower prevalence rates were also reported in other countries. For instance, only 0.8% and 3% prevalence rates were reported in Turkey (31) and Greece (32), respectively, but Japan and Lebanon reported prevalence rates of 4.7% and 10.5%, respectively (6, 26). This is not surprising because variations in the prevalence rates of MLSB phenotype have been recognized due to the differences in geographic regions, local institutions, type of strains, and local antibiotic policies (6, 8, 9).
The low prevalence rate of MSLB resistance in the present study could probably be explained by the judicious use of antibiotics in the primary care setting in Malaysia. This is supported by the fact that all MSSA isolates from healthy carriers were sensitive to antibiotics in the present study. In Malaysia, simply buying antibiotics over the counter is not possible; a prescription from a physician is required for the purchase. In addition, antibiotic stewardship has been implemented in most Malaysian hospitals. Thus, the use of antibiotics for in-patients is closely monitored.
Surprisingly, no cMLSB phenotype was detected in the present study. A study in Japan demonstrated that the iMLSB phenotype was more prevalent than the cMLSB phenotype in their MSSA isolates (94% versus 1.3%). In their study, erythromycin was not used as the first-line treatment in the community setting, which could explain the difference between MSLB phenotypes (6). However, other studies have reported cMLSB as the predominant phenotype (31).
With regard to the distribution of MLSB resistance genes, the ermC gene was the most predominant gene found despite the low number of iMLSB strains in the present study. No other erm genes were detected. Our finding was higher than reported in other countries. In Turkey and Japan, 52.9% and 41.8% of their iMLSB strains predominantly contained the ermC gene, respectively (6, 31). However, the ermA gene was more predominantly detected than ermC gene (58.3% versus 20%) in Greece (32). The iMLSB phenotype has commonly been associated with the ermC gene (6). The concern of these findings in the present study cannot be ignored because the ermC gene can be easily transferred by plasmids to other species. Thus, continued antibiotic surveillance is warranted.
Only one MS phenotype with the msrA gene was detected in the present study. The msrA or msrB genes are commonly detected in MS phenotype strains. Staphylococcus aureus with the MS phenotype should be properly recognized because the mechanism of its resistance is due to an altered efflux system, and this bacterium is still susceptible to clindamycin despite its resistance to erythromycin (14). Thus, the use of clindamycin is still appropriate for treatment, especially for those who are allergic to penicillin.
Nonetheless, our study has several limitations. First, our study might not be representative of local MSSA colonizers because only a small proportion of healthy carriers were included in the study. Furthermore, the MSSA clinical isolates were only collected at one healthcare center; community-acquired MSSA isolates should have been included as well. Finally, other genes, such as ermF and ermY, were not included in the present study. These newly recognized genes are also responsible for MLSB resistance.
Judicious use of antibiotics in hospital and community settings is very important to control the emergence of antibiotic resistance isolates. However, the presence of ermC gene, despite the low frequency of iMLSB phenotype isolates in the present study, cannot be ignored because failure to recognize this resistance phenotype would affect therapeutic management and promote bacterial resistance in future. In addition, labelling all erythromycin-resistant S. aureus isolates as clindamycin resistant would prevent the use of clindamycin in infections caused by clindamycin susceptible isolates. The existing antibiotic surveillance system is still necessary to control any emergence of resistance isolates so that targeted therapy and effective control can be implemented accordingly.