1. Background
Streptococcus agalactiae is an important cause of pneumonia, sepsis, and meningitis in low-immunity individuals (1). Specifically, S. agalactiae infection has been associated with significant rates of prenatal and neonatal infections worldwide (2). Maternal S. agalactiae colonization is associated with urinary tract infections, endometritis, chorioamnionitis, premature delivery, and intra-uterine death (3). Of pregnant women colonized with S. agalactiae, 40% - 70% may transfer S. agalactiae to their newborns; 1% - 3% of newborns develop septicemia, meningitis, pneumonia, and early neonatal sepsis with a 10% - 20% mortality rate (4).
The US centers for disease control and prevention (CDC) recommended screening of all pregnant women for S. agalactiae between 35 and 37 weeks of gestation (5). As a consequence of screening, there was a 75% reduction in neonatal infections caused by S. agalactiae. The CDC recommends a standard identification method based on culture results; however, this method has a slow turnaround time, requiring 36 - 72 hours for processing before the results are available and requires experienced technicians (6). A rapid, accurate, sensitive detection of S. agalactiae in the early stage of infection is a major breakthrough in clinical diagnosis. Molecular methods may also be helpful (7, 8).
Loop-mediated isothermal amplification (LAMP) is well-established and documented and has been verified to be effective in bacterial identification. Loop-mediated isothermal amplification is a novel nucleic acid amplification method that amplifies DNA with high specificity, efficiency, and rapidity. LAMP is performed under 60 - 65°C isothermal conditions using a set of four specifically designed primers that recognize six distinct regions of the target gene and Bst DNA polymerase with strand displacement activity (9-11). The reaction results in large amounts of turbid amplification products that can be easily detected by naked eyes without magnification and no gel electrophoresis is required. Therefore, the LAMP assay has become a powerful tool for the rapid diagnosis of infectious diseases or specific genes of microorganisms (12, 13).
Therefore, a simple and rapid diagnostic method for detection of S. agalactiae using LAMP assay is reported here.
2. Methods
2.1. Bacterial Strains
A standard strain of S. agalactiae ATCC 13813 was used to establish and optimize the LAMP reaction. Clinical isolates of S. agalactiae, S. pyogenic, S. viridans, Group F Streptococcus, S. pneumoniae, Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa from our hospital were used to evaluate the specificity of the LAMP assay using standard biochemical tests and the MicroScan WalkAway 96 system (siemens healthcare diagnostics Inc.,Tarrytown, NY, USA). Also, 40 clinical specimens infected with bacteria were included in this study to evaluate the application of the LAMP assay in the clinical settings.
2.2. DNA Isolation
For genomic DNA extraction, all the bacteria were cultured on Columbia blood agar (Autobio, Zhenzhou, China) at 37°C for 20 hours. Bacterial genomic DNA was prepared by boiling the specimens in distilled water (DW) for 10 minutes. Then, the lysate was centrifuged at 12,000 × g for 2 minutes, and the supernatant was transferred to a new tube and designated as “genomic DNA”. After measuring the concentration using Nanodrop 2000 (Bio-Rad, Hercules, CA, USA), all the DNA was diluted to 100 ng/μL to be used as a template for LAMP and PCR amplification in the following experiments.
2.3. Design of LAMP Primers
In order to identify S. agalactiae, the species-specific cAMP gene was selected as the target gene (GenBank accession No. NC-004368). Oligonucleotide primers for LAMP were designed using Primer Explorer V4 software (http: //primerexplorer.jp/e/), and synthesized commercially by invitrogen biotech (Invitrogen, Carlsbad, CA, USA). Primers (F3, forward outer primer; B3, backward outer primer; FIP, forward inner primer; BIP, backward inner primer) were specifically designed for the LAMP reaction to target six distinct regions (Tsugunori, 2000).
2.4. LAMP Reaction and Primer Screening
The LAMP reaction was carried out in a 25 μL reaction mixture according to the manufacturers’ instructions (Eiken China Co., Ltd., Shanghai, China). 1 μL fluorescent detection reagent (Eiken) was added to help in interpreting the results. Negative and positive controls were established according to the kit instructions. After primers were added, the reaction tubes were incubated at 63°C for 60 minutes in the loopamp real-time turbidimeter (LA-320 C; Eiken Chemical Co., Ltd. Tokyo, Japan) to screen the optimal primer set.
2.5. Results Interpretation
The LAMP products were detected based on color change. The color of the reaction mixture turned green in the presence of LAMP amplification, while the color remained orange in negative amplification tubes.
2.6. S. agalactiae Detection by PCR
Polymerase chain reaction (PCR) assays were performed using the same genomic DNA as templates. Outer primers (F3 and B3) were used as PCR forward and reverse primers, allowing the amplification of a sequence of 202 bp. The PCRs were carried out in a 25 μL reaction mixture consisting of 12.5 μL of 2 × taq mastermix (Cwbiotech, Beijing, China), 10 μM F3 and B3 primers, 2 μL of DNA, and 8.5 μL of DW. The reaction conditions were carried out at 94°C for 2 minutes, followed by 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes, and a final extension at 72°C for 2 minutes. The PCR products were then analyzed on a 1% agarose gel (Biowest, Nuaille, France) by electrophoresis. DW was used as a negative control.
2.7. Sensitivity of LAMP Assays
In order to determine the sensitivity of the LAMP method using the screened primer set for identification of S. agalactiae cAMP gene, the S. agalactiae ATCC 13818 culture was serially diluted 10-fold with DW, ranging from 105 to 1 CFU/mL. Then the genomic DNA was extracted and LAMP reactions were set up using the aforementioned DNA at 63°C. For a change, the reaction tubes were incubated in a thermostatic water bath instead of the Loopamp Real-time Turbidimeter. For comparison, PCR assays were performed using the same diluted genomic DNA as templates. S. agalactiae genomic DNA was diluted 10-fold incrementally, ranging from 0.001 to 100 ng/μL, undergoing the same amplification procedure.
2.8. Specificity of LAMP Identify
In order to validate the specificity of the LAMP assays, six species of bacteria that were likely associated with S. agalactiae in human infections, but without cAMP genes, were tested including S. pyogenic, S. viridans, S. constelltus, group F Streptococcus, S. pneumoniae, and Staphylococcus aureus. The genomic DNA was extracted and the LAMP reactions were carried out as discussed above. The PCR method was used for comparison.
2.9. Application of LAMP Assays on Clinical Isolates
The above LAMP reaction was carried out using pure culture lysates. For rapid diagnosis of S. agalactiae in the clinical settings, this LAMP assay must be able to detect the S. agalactiae gene in clinical specimens. Forty vaginal swabs were collected from hospitalized patients. Swabs were first placed into test tubes with 1 mL of DW and were shaken adequately. Then, the swabs were discarded and the remaining liquid was centrifuged at 10,000 × g for 2 minutes. The sediments were collected and DNA extraction performed as discussed above. All of the DNA was used as a template for LAMP and PCR. The results of the clinical diagnosis were used as a standard
3. Results
3.1. Optimal LAMP Primer for S. agalactiae Detection
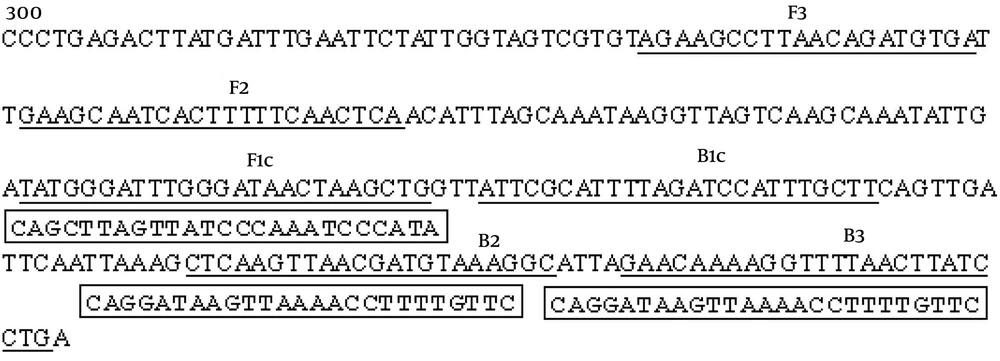
We initially screened one set of optimal LAMP primers for S. agalactiae detection from five sets of primers in which S. agalactiae DNA was detected by the LAMP reaction in 39 minutes. The positions and sequences of the optimal primer set are shown in Figure 1 and Table 1. The nucleotide sequence of the cAMP gene (GenBank accession No.NC-004368) is shown, and the number indicates the nucleotide positions. The locations of the primer binding sequences are underlined. FIP = F1c + F2, BIP = B1c+ B2
| Primer | Sequences (5’ - 3’) | Length |
|---|---|---|
| F3 | AGAAGCCTTAACAGATGTGA | 20 bp |
| B3 | CAGGATAAGTTAAAACCTTTTGTTC | 25 bp |
| FIP | CAGCTTAGTTATCCCAAATCCCATAGAAGCAATCACTTTTTCAACTCA | 48 bp |
| BIP | ATTCGCATTTTAGATCCATTTGCTTGCCTTTACATCGTTAACTTGAG | 47 bp |
Primer Sequences
3.2. Detection of LAMP Products
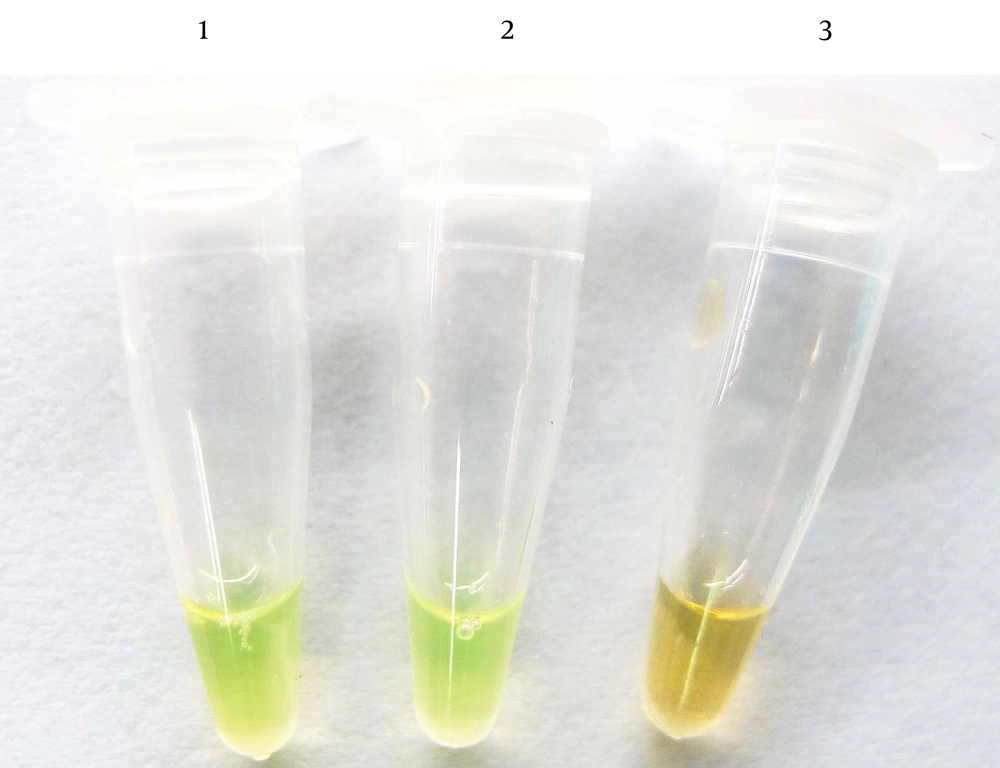
Consistent with the DNA amplification kit manual, Figure 2 shows that the liquid color of positive reactions changed from orange to green, while negative reactions remained orange. Results observed without magnification. 1: S. agalactiae; 2: positive control; 3: negative control.
3.3. Sensitivity of S. agalactiae Identification by the LAMP Method
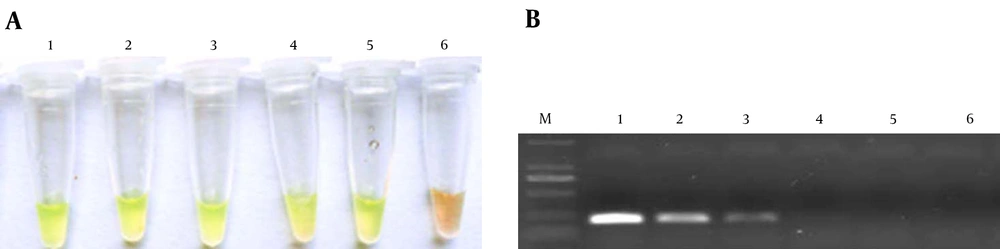
As shown in Figure 3, the limit of detection (LOD) of the LAMP assay for genomic DNA was approximately 0.01 ng/μL of DNA. For S. agalactiae, the LOD was approximately 10 CFU/mL (results not shown). For PCR the concentration limits were 1 ng/μL of genomic DNA and 100 CFU/mL of S. agalactiae. Compared with PCR, LAMP has 10 - 100-fold more sensitivity.
3.4. Specificity of the LAMP Method
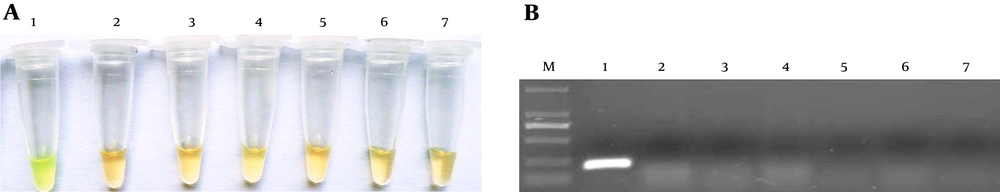
Figure 4 shows that the selected primer set only amplified the cAMP gene of S. agalactiae, but no amplification was found using the specific primer set for other bacterial species; thus, the LAMP method using this primer set was specific for S. agalactiae identification. The PCR method using F3 and B3 as primer set for detection of the S. agalactiae cAMP gene had the same specificity here.
3.5. Detection of S. agalactiae in the Clinical Settings
Forty clinical samples were examined by LAMP method. The results showed nine positive and 31 negative samples, which is consistent with culture-based identification and clinical diagnosis. Only three positive samples were detected by the PCR method because of the low sensitivity. Advantages of LAMP over PCR method were shown in Table 2.
| LAMP | PCR | |
|---|---|---|
| Sensitivity | 0.01 ng/μL | 1 ng/μL |
| Specificity | 100% | 85% |
| Time cost | < 1 h | 1.5 - 3 h |
| Convenient | ||
| Instrument | Water bath | PCR Amplifier |
| Judge of results | Naked eyes | electrophoresis |
Differences Between LAMPand PCR Method
4. Discussion
Streptococcus agalactiae is recognized as a lethal pathogen in neonates worldwide. Its infections severely affect pregnant women and immunosuppressed adults with substantial morbidity and mortality. A 17-year study showed that there was a remarkable increase in S. agalactiae infections over time (14). It has been verified that S. agalactiae screening among pregnant women significantly decreases neonatal infections (5). The traditional identification methods for S. agalactiae are complex and time-consuming (6). So, a rapid, accurate, sensitive, and easily-manipulated detection method for S. agalactiae is needed in the clinical settings.
In the current study, the LAMP method was used for detection of S. agalactiae. Compared with the traditional culture detection or PCR method,there are several advantagessuch as timesaving. Using the selected primer set, the species-specific cAMP gene of S. agalactiae was amplified and detected without magnification in 40 minutes. While culture detection requires 36 - 72 hours (6, 15) and antigen detection or PCR-based assay need over two hours (7, 16-18). Secondly, LAMP method has high sensitivity for detection of S. agalactiae. The LOD of genomic DNA of S. agalactiae was 0.01 ng/μL, which is equivalent to 1 - 10 CFU/mL of S. agalactiae. The LAMP method was 10 to 100-fold more sensitive than PCR. Thirdly, The LAMP detection method has high accuracy. Here, all LAMP detection results were 100% consistent with the culture-based detection and clinical diagnosis. PCR only detected S. agalactiae in 85% of the clinical specimens. Fourthly, LAMP method was very convenient and easy to use. In terms of ease of use, firstly LAMP reaction could be performed in a normal water bath in lieu of using any special equipment. Secondly, the results of the LAMP could be assessed by naked eyes without magnification which is the most important feature of this assay.
In a word, LAMP method is a clinically useful molecular diagnostic test, especially for the rapid screening of S. agalactiae. Moreover, the rapid detection of S. agalactiae will be helpful and LAMP can be used as a universal screening method among pregnant women which subsequently can result in reduction of infection between newborns