1. Background
Helicobacter pylori, previously known as Campylobacter, is a microaerophilic Gram negative bacterium that is able to colonize the gastric mucosa layer (1). The high prevalence of H. pylori infection is a major problem, especially in developing countries (2, 3). The infection is most often acquired during early childhood, and many studies have shown that transmission is within families (4, 5). Although all strains cause gastritis to varying degrees, only some strains are significantly associated with an increased risk of gastroduodenal diseases, such as peptic ulcer disease and gastric cancer (6). Therefore, to determine strains associated with gastric cancer (GC) and peptic ulcer disease (PUD), as well as to understand the role of H. pylori infection in a wide range of extragastric diseases, effective diagnostic methods are mandatory (7). Several methods are available for the identification of H. pylori that can be classified as invasive techniques requiring endoscopy, such as Campylobacter-like organism test (CLO test), culturing, histological examination and PCR, or non-invasive techniques such as serology, which is specifically recommended for patients with dyspeptic symptoms in primary care (8). Among these tests, PCR is a very sensitive and specific technique capable of determining different strains of H. pylori (9, 10).
None of these diagnostic methods are flawless, and studies have shown that a combination of at least two diagnostic tests (usually biopsy-based tests), gives more accurate diagnosis (9). The main advantage of PCR assay is that it can be performed with the same biopsy sample used for CLO test and the biopsy sample requires no rigorous transport conditions (11, 12). A reduction in gastric acid secretion (hypochlorhydria) leads to the erratic distribution of H. pylori in the stomach, resulting in flawed concordance rates between different diagnostic methods (10, 13). It also allows the colonization by other urease-producing microorganisms (14-16). Achlorhydria may occur in patients with previous gastric surgery or, patients treated with long-term antibiotics or antisecretory drugs, especially proton pump inhibitors (PPI) (10, 13).
In our previous study, we found the prevalence of 72% for H. pylori infection in East Azerbaijan, Iran; almost 100% of the strains carried the vacA d region; and a significant association was observed between the vacA d1 genotype and gastric adenocarcinoma and PUD (17). Several studies have suggested that the elimination of H. pylori may increase the risk of diseases such as esophageal adenocarcinoma (18). Considering the high incidence of gastric (ASR: 37.6) and esophageal cancer (ASR: 24.1) compared to other gastrointestinal tract cancers in this area (11), the identification of cancer-related strains before eradication is important.
2. Objectives
The present study aimed to determine cancer-related strains by PCR, using the gene predictor of gastric adenocarcinoma and PUD (vacA d region) and 16S rDNA gene, in patients with gastritis and PUD. Since none of H.pylori detection methods are perfect, the PCR assay was performed with the same biopsy sample used for CLO test.
3. Methods
3.1. Patients
The specimens used for the study were collected from patients over 16 years old who received diagnostic endoscopy at Imam Reza hospital, Tabriz, Iran. Demographic data, such as language, nationality, gender and family history of gastric cancer or gastric ulcer were collected from all patients. Patients treated with long-term antibiotics or antisecretory drugs, especially PPI, as well as patients having previous history of PUD, GC, endoscopic dissection and gastrectomy were excluded (12, 19). Patients with gastric cancer were also excluded after a clinical examination. Antral gastric biopsies were collected from 30 patients with diseases of gastritis and PUD. Informed consent was obtained from all the recruited patients, and approval for sample collection was granted by the ethics committee of the hospital. The sample consisted of 12 males and 18 females with mean age of 42 years.
3.2. Collection of Samples
A total of 30 specimens were collected from patients with gastritis and PUD. One biopsy sample from each patient was collected and used for both tests (CLO test and PCR assay). The CLO test was first performed on the biopsy specimen, followed by DNA extraction and PCR assay.
3.3. Campylobacter-Like Organism Test
One piece of antral biopsy was introduced with a sterile needle into a urease tube. The test tube was incubated at 35°C in ambient air for an hour and a half. Once the color changed from yellow to magenta indicating the presence of H. pylori, specimens were removed from the urease tube with a sterile needle and then transferred to sterile tubes and frozen at -80°C until processing. In order to prevent confounding caused by other urease-positive bacteria such as streptococci and staphylococci (which have the ability to yield a lower amount of urease), the specimens were removed from CLO test tubes after a short-time detection (< 2 hours) (7).
3.4. DNA Extraction and PCR
First, DNA extraction was performed by crushing the biopsy specimens using two slides, and then by using the DNGTM-Plus kit (CinnaGen Co, Iran) in accordance with the manufacturer’s protocol. In this study, genotyping of the 16S rDNA gene and the vacA d region (to identify strains associated with GC and PUD) was accomplished by PCR assay. All primer sets were selected from published literature (20, 21). The primers (CinnaGen Co, Iran) 5’ -GCAATCAGCGTCAGTAATGTTC- 3’ (forward primer HP1) and 5’-GCTAAGAGATCAGCCTATGTCC- 3’ (reverse primer HP2) were used to amplify a fragment of 519 base pairs (bp) of 16S rDNA gene, and the primers (Macrogen Co, South Korea) 5’ -ACTAATATTGGCACACTGGATTTG- 3’ (forward primer VAS-5F) and 5’ -CTCGCTTGATTGGACAGATTG- 3’ (reverse primer VAGF-R) were used to amplify 367-379 pb and 298 pb products from the vacA d1 and d2 alleles, respectively. Briefly, PCRs were performed in a total volume of 25 μL using a commercially available kit (CinnaGen Co, Iran), under the following conditions for the vacA d1 and 16S rDNA amplification: 95°C for 5 minutes; then 37 cycles of 95°C for 45 seconds, 53°C for 60 seconds, and 72°C for 30 seconds; and 72°C for 5 minutes. Finally, PCR products were examined by gel electrophoresis on 1.2% agarose gel, and analyzed in a UV transilluminator after staining with ethidium bromide.
4. Results
Twenty-four (80%) of the biopsy samples showed positive results on PCR assay (vacA d and 16S rDNA genes) from which, 19 (79.2%) were classified as gastritis, and 5 (20.8%) as peptic ulcer. This study showed that the frequencies of the vacA d1 and d2 were 20.8% and 79.2%, respectively. The frequency of the allele d1 in H. pylori isolates was higher in patients with peptic ulcer (60%) than those with gastritis (10.53%) (P = 0.002). One of the patients with gastritis carrying the vacA d1- positive strains had a history of gastric cancer in first-degree relatives. However, no family history of gastric cancer or gastric ulcer was found in the patients with strains carrying the vacA d2 genotype.
The same biopsy samples were used for both the CLO test and PCR assay. Results also revealed that out of 22 (73.3%) CLO test-positive samples, 100% were also positive on the PCR assay; while out of 8 (26.7%) CLO test-negative samples, 6 (75.00%) were also negative on the PCR assay (Table 1).
| Samples | N = 30 |
|---|---|
| CLO test-negative and PCR- negative | 6 |
| CLO test-positive and PCR- positive | 22 |
| CLO test-negative and PCR- positive | 2 |
In this study, the frequency of the false negative CLO test results was 2/30 (8.3%). However, no false-positive CLO test results were found.
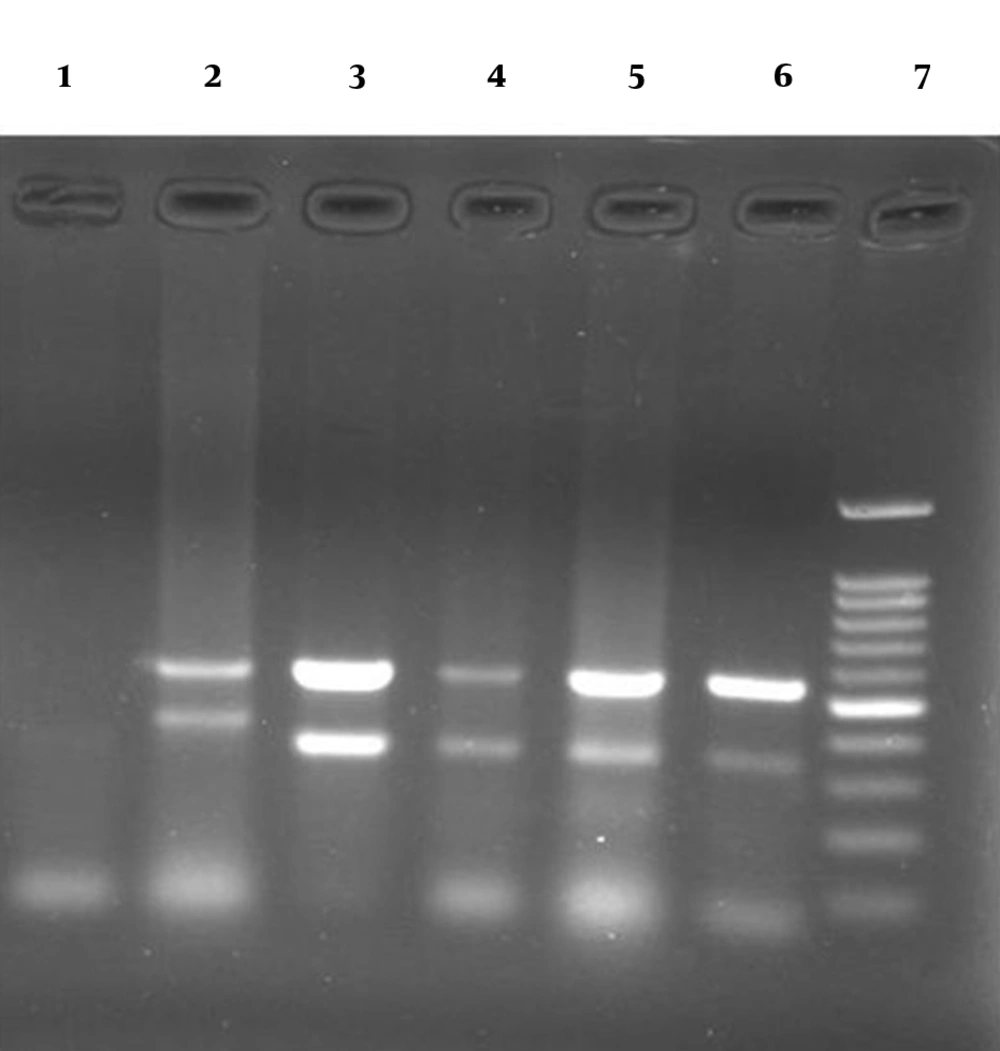
Agarose gel (1.2%) containing PCR products from allele d1 (367-379 pb) and allele d2 (298 pb) of vacA gene and 16S rDNA gene (519 bp): lane 2: positive control (Strains J99 and Tx30a) representing the allele d1, lanes 3-6: clinical positive strains representing the allele d2, lane 1: negative control and lane 7: 100-bp DNA ladder
5. Discussion
Since 1994, when H. pylori infection was recognized as a definite class I human carcinogen, numerous methods have been developed to detect and eradicate H. pylori infection (8, 22). The need for appropriate methods (especially a combination of two or more biopsy-based tests) that accurately detect strains associated with GC and PUD is growing; in this regard, there is an increasing focus on the eradication of H. pylori infection associated with a range of extragastric diseases, especially in countries where gastric cancer is common (9, 17, 18). The aim of this study was to determine cancer-related strains by PCR assay and CLO test in patients with gastritis and PUD. The highly sensitive and specific PCR assay is increasingly used in clinical studies owing to its capability to detect different strains of H. pylori (23). In the present study, gene targets for PCR methods included the highly conserved 16S rDNA gene (with a high sensitivity), which has been widely used (24) and the vacA d region that is highly sensitive and specific in predicting gastric adenocarcinoma and PUD in different geographic regions (17, 21, 25). In this study, the 16S rDNA and vacA d PCRs were able to detect a high percentage (79.1%) of H. pylori infection in patients with PUD and gastritis, a finding consistent with results of previous studies (17, 21, 24, 26). Furthermore, the existence of strains with the vacA d1 genotype (a predictor of gastric adenocarcinoma and PUD) in gastritis patient with a family history of gastric cancer confirms the notion of intrafamilial transmission of the infection (5).
Although CLO test is highly specific, it requires a high H. pylori density for detection, and false positive results could occur in patients with achlorhydria (14-16). On the other hand, studies have shown that the achlorhydria may not affect the rate of H. pylori detection by PCR assay, because the genetic material of a bacterium remains unchanged over time (27). In a study conducted by Tin-Tsan Lin et al. (1996) on 4 (6.5%) cases with false-positive results, 3 (75%) had been treated with long-term H2 blockers. In the present study, patients with suspected achlorhydria were first excluded and then, any possible false result on the CLO test was examined. In agreement with the findings from other studies, no false-positive CLO test results were found for biopsy specimens in this study (28, 29). There were 2 (8.3%) false-negative CLO test results in this study and also, half of the 8.3% cases (2 patients) with false negative CLO test results had PUD.
Electrophoresis revealed a significantly higher density of PCR products for all the 24 cases with positive PCR and negative CLO test results. This study suggests that the vacA d PCR to determine strains associated with GC and PUD can be considered very useful tool in diagnosis, treatment, and follow-up of patients with gastritis and PUD. Consistent with other studies, the findings of the present study imply that factors causing achlorhydria can lead to false positive CLO test results (14-16, 30). The use of the same biopsy sample for both the CLO test and PCR assay could lead to a decrease in false negative CLO test results (29, 31). The presence of a higher density of PCR products in the electrophoresis of samples collected from CLO test requires further study.