1. Background
Enterococcus faecium is an important nosocomial pathogen. Notaexpression of nitroreductasebly, it has developed resistance to multiple antibiotics, including vancomycin (1). The emergence of vancomycin resistance is most commonly found in E. faecium, and the incidence of E. faecium infections is increasing; thus, treatment of these infections is difficult (2). Nitrofurantoin belongs to a group of compounds characterized by the presence of one or more nitro-groups on a nitroaromatic or nitroheterocyclic backbone. It is taken orally, rapidly absorbed and excreted in the urine to generate high therapeutic concentrations (3).
Nitrofurantoin is active against E. faecium and E. faecalis, and it retains its activity against vanA- and vanB-positive isolates (4). Although the specific mode of action of nitrofurantoin is still unknown, strains resistant and susceptible to nitrofurantoin differ in their ability to reduce various compounds (3). Stein et al. (5) reported that the mutants lacking nitroreductases are more resistant to nitrofurantoin due to their decreased activity.
Du et al. (6) obtained a E. faecium Rifr mutant in a previous study. Rifr mutants were isolated spontaneously from rifampin plates. We determined the growth rate and reactive oxygen species (ROS) levels to further characterize this mutant (B42-R7, rpoB H489D). We also compared the proteomic profiles of E. faecium B42 and its Rifr mutant B42-R7. The induced nitroreductase in B42-R7 may explain why B42-R7 became susceptible to nitrofurantoin.
2. Methods
2.1. Bacterial Strains, Media and Antibiotics
Restriction enzymes, T4 ligase, and Taq DNA polymerase were purchased from TaKaRa (Otsu, Shiga, Japan). All Enterococci cultures were grown at 37°C in Brain-Heart Infusion (BHI) broth and agar (Oxoid, Basingstoke, UK) (Appendix 1 in Supplementary File). All Escherichia coli cultures were grown at 37°C in Luria-Bertani (LB) broth and agar (Oxoid, Basingstoke, UK). Nitrofurantoin MICs were determined by Etest (bioMerieux, France) on Mueller-Hinton (MH) agar (Oxoid, Basingstoke, UK).
2.2. Double Time Measurement
Growth was measured using two methods. The first method was performed as described previously (7). Briefly, 500 μL overnight culture of each strain was transferred to 50 mL of BHI. The culture was grown at 37°C while shaking at 250 rpm. The dilutions were spread onto BHI agar plates after 2 hours (t1) and 4 hours (t2). The plates were incubated at 37°C, and the number of colony forming units (CFU) was determined. The doubling time (g) was calculated from g = ln2 / ((log10 N2 - log10 N1) 2.303/Δt), where N1 is CFU per mL at t1, and N2 is CFU per mL at t2. The second method used the Bioscreen C reader (Oy Growth Curves Ab Ltd., Finland) to determine the growth rate of the strains in BHI broth at 37°C (8). Six independent cultures per strain were grown overnight, diluted 1:1000 and aliquoted into a Bioscreen C plate in triplicate. The growth rate was estimated by an R script from an OD600 interval between 0.02 and 0.08.
2.3. ROS Measurement
Overnight-cultured bacteria were diluted 1:100 into 1 mL fresh BHI culture. The bacteria were harvested and washed with PBS after the dilutions and were incubated in 37°C for 2 hours. Then, bacteria were incubated with dichlorofluorescein-diacetate (DCFH-DA, 1 µM) for 30 minutes at 37°C and washed with PBS. The fluorescence of the solution was measured using a spectrofluorometer (excitation, 500 nm; emission, 530 nm) (9). DCFH-DA was purchased from Molecular Probes, Inc. (Eugene, OR).
2.4. Preparation of Whole Cellular Extracts
Overnight cultures of E. faecium B42 and B42-R7 were diluted 1:100 in BHI and grown to an OD600 of 1.0 at 37°C at 250 rpm. Then, the bacteria were centrifuged for 5 min at 6000 × g and washed three times with PBS. These experiments were repeated three times (10).
Cell pellets were resuspended in 1 mL of lysis buffer (9.5 M urea, 65 mM DTT, 4% (w/v) CHAPS and 0.2% IPG buffer) containing a protease inhibitor cocktail. The cells were lysed for 30 minutes at room temperature. Then, the cells were sonicated for 5 minutes (cycles of 10 seconds of sonication with 15 seconds intervals) on ice. The cell lysates were centrifuged for 30 minutes at 12,000 × g. Then, the supernatants were collected, and the protein concentrations were determined using Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA, USA). The prepared samples were stored at -80°C (10).
2.5. Two-Dimensional Electrophoresis (2-DE)
For 2-DE, 100 μg and 400 μg of proteins were loaded onto analytical and preparative gels, respectively. The Ettan IPGphor Isoelectric Focusing System (Amersham Biosciences, Buckinghamshire, UK) and pH 3-10 immobilized pH gradient (IPG) strips (13 cm, nonlinear, Amersham Biosciences, Buckinghamshire, UK) were used for isoelectric focusing (IEF). The IPG strips were rehydrated for 12 hours in 250 μL of rehydration buffer containing the protein samples. IEF was performed in four steps: 30 V for 12 hours, 500 V for 1 hour, 1000 V for 1 hour, 8000 V for 8 hours, and 500 V for 5 hours. The strips were then subjected to the second-dimensional electrophoresis after transfer onto 12.5% SDS-polyacrylamide gels. Electrophoresis was performed using the Hofer SE 600 system (Amersham Biosciences, Buckinghamshire, UK) at 15 mA per gel for 30 minutes, followed by 30 mA per gel until the bromophenol blue reached the end of the gel.
Protein spots in the analytical gels were visualized by silver staining. The preparative gels were stained using a modified silver staining method compatible with subsequent mass spectrometric analysis (11). The stained gels were scanned using UMax Powerlook 2110XL (UMax), and image analysis was performed using ImageMaster 2D Platinum (GE Healthcare, Sweden). Each paired spot was manually verified to ensure a high level of reproducibility between normalized spot volumes of gels produced in triplicate data. The overlapping measures ratio was chosen to identify protein expression changes, and proteins with a 1.5-fold or greater overlap ratio threshold filtering were considered differentially expressed. Three replicates were performed for each sample.
2.6. 2D Digestion
Protein spots were cut from the preparative gels, destained for 20 minutes in 30 mM potassium ferricyanide/100 mM sodium thiosulfate (1:1 v/v) and washed with Milli-Q water until the gels were destained. Each spot was lyophilized and digested overnight in 12.5 ng/mL trypsin. The peptides were extracted three times with 60% ACN/0.1% TFA. The extracts were pooled and dried completely by a vacuum centrifuge (12).
2.7. MALDI-TOF/TOF MS Analysis
The dry peptide samples were reconstituted in 2 μL 20% ACN, spotted on a 384-well Opti-TOF stainless steel plate, and covered with 5 mg/ml cyano-4-hydroxycinnamic acid (CHCA) in 50% ACN and 0.1% trifluoroacetic acid (TFA) before being dried. MS and MS/MS data for protein identification were obtained using a MALDI-TOF-TOF instrument (4800 proteomics analyzer; Applied Biosystems, Framingham, MA). Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems, Framingham, MA). The MS spectra were recorded in reflector mode in a mass range from 800 to 4000 Da with a focus mass of 2000 Da. MS was used with a CalMix5 standard to calibrate the instrument (ABI 4700 Calibration Mixture). For one major MS spectrum, 25 sub-spectra with 125 shots per sub-spectrum were accumulated using a random search pattern. For MS calibration, autolysis peaks of trypsin ([M + H]+ 842.5100 and 2,211.1046) were used as internal calibrators. Additionally, up to 10 of the most intense ion signals were selected as precursors for MS/MS acquisition, excluding the trypsin autolysis peaks and the matrix ion signals. In MS/MS positive ion mode, for one major MS spectrum, 50 sub-spectra with 50 shots per sub-spectrum were accumulated using a random search pattern. Collision energy was 2 kV, collision gas was air, and default calibration was set by using the Glu1-Fibrino-peptide B ([M + H]+ 1,570.6696) spotted onto Cal 7 positions of the MALDI target.
Combined peptide mass fingerprinting PMF and MS/MS queries were performed using the MASCOT search engine 2.2 (Matrix Science, Ltd.) embedded into GPS-Explorer Software 3.6 (Applied Biosystems) on the NCBI database (downloaded 2011-12-29) with the following parameter settings: 100 ppm mass accuracy, trypsin cleavage one missed cleavage allowed, carbamidomethylation set as fixed modification, oxidation of methionine was allowed as variable modification, MS/MS fragment tolerance was set to 0.4 Da. A GPS Explorer protein confidence index ≥ 95% was used for further manual validation.
2.8. Complementation Experiments
The E. faecium B42 nitroreductase gene was PCR-amplified from genomic wild-type DNA and ligated into the E. coli-Enterococcus shuttle vector pDL276 to generate the plasmid pDL276-NR. The pDL276-NR plasmid and pDL276 plasmid were transformed into E. coli DH5α cells, and transformants were selected for their ability to grow on kanamycin. Transformation was confirmed by PCR amplification and sequencing of the kanaR gene in pDL276. Nitrofurantoin MICs of transformants were determined by Etest on Mueller-Hinton (MH) agar (Oxoid, Basingstoke, UK).
The nitroreductase gene was also sub-cloned into pBSU101 to generate the plasmid pBSU101-NR. Both pBSU101 and pBSU101-NR were transformed into E. faecium ATCC 35667. Nitrofurantoin MICs of DH5α cells harboring pBSU101 or pBSU101-NR were determined by Etest on BHI agar.
3. Results
3.1. rpoB H489D Had a Lower Growth Rate and Higher ROS Levels
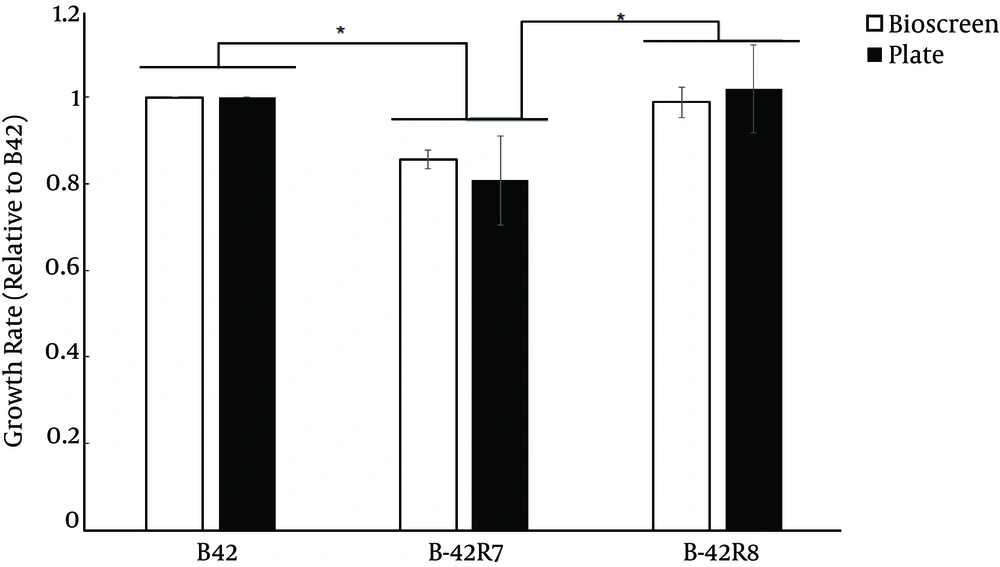
To determine whether the mutation in rpoB resulted in a fitness cost, we measured the doubling time for exponentially growing cells using two methods and assessed the wild type, B42-R7 and B42-R8. B42-R8 showed similar growth rates as the wild type (Figure 1). However, B42-R7 had a lower growth rate than the wild type. A similar trend was also reported for rpoB mutants of Deinococcus radiodurans (7).
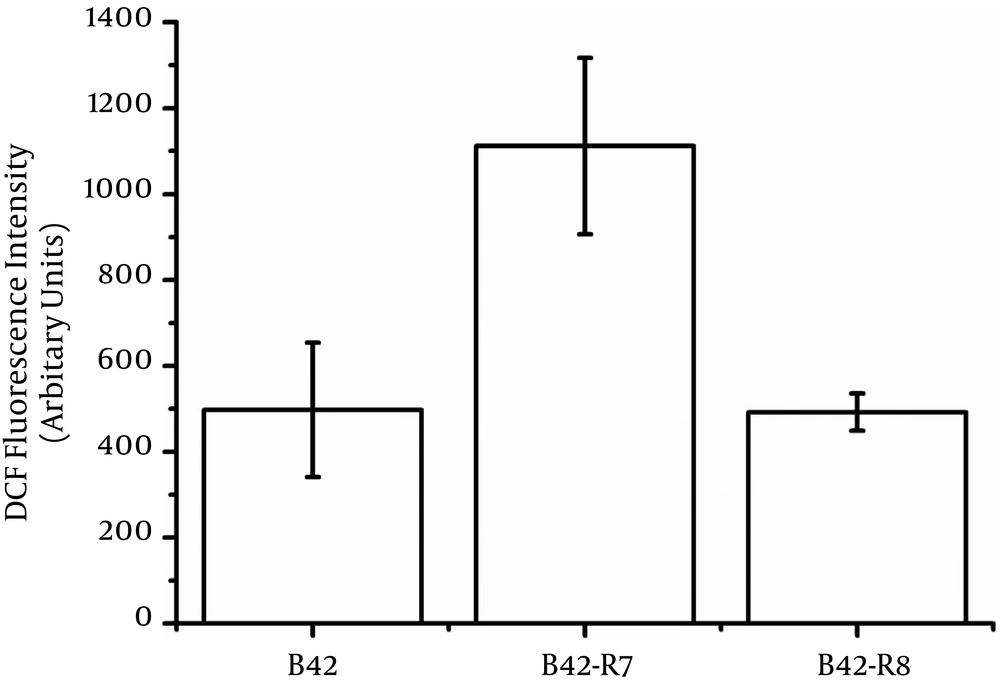
We also measured the intracellular ROS levels of wild type and mutants. B42-R7 had higher intracellular ROS levels than those of the wild type (Figure 2). The rpoB H489P mutation did not induce intracellular ROS.
3.2. Two-Dimensional Gel Analysis of Differentially Expressed Proteins of E. faecium and Its Rifr RpoB H489D Mutants
In order to investigate the proteomic response and possible proteins involved in nitrofurantoin resistance, we compared the proteomic profiles of E. faecium B42 and its Rifr mutant B42-R7. We identified a total of 63 spots with altered expression, including 24 spots that were up-regulated and 39 spots that were down-regulated (Appendix 2 in Supplementary File). After analysis using a 1.5 fold-change filter, 6 spots were up-regulated, and 15 spots were down-regulated. After the 6 and 15 spots were identified by MALDI-TOF/TOF MS/MS mass spectrometry, we obtained 4 and 11 proteins, respectively (Table 1 Appendix 3 in Supplementary File). These proteins included (1) proteins with a known nitrofurantoin resistance function; (2) metabolism-related proteins, which are necessary for growth, especially those related to glycolysis, energy production and conversion, and nucleotide transport and metabolism; (3) proteins related to translation and transcription; (4) a hypothetical protein: signal peptide.
| Spot no | NCBI GI identifier | COG | Protein description | Theor. Mass | Score | Sequence Coverage% | Theor. pI | Location | Gene | Fold change | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Down-Regulated in B42-R7 | |||||||||||
| 649 | 227552066 | G | glyceraldehyde-3-phosphate dehydrogenase | 35912.2 | 428 | 60 | 5.26 | C | gap-2 | - 4.87057 | 0.967097 |
| 770 | 227551650 | E | 3-dehydroquinate synthase | 38490.4 | 170 | 18 | 5.56 | C | aroB | - 2.70866 | 0.708657 |
| 1255 | 257867607 | TK | transcriptional regulator | 26393.8 | 176 | 25 | 5.12 | C | - 10.5638 | 1.053339 | |
| 476 | 69247805 | C | Glutathione reductase, animal and bacteria | 49718.3 | 699 | 56 | 5.55 | C | - 2.06685 | 0.172891 | |
| 269 | 69249876 | G | Glyceraldehyde-3-phosphate dehydrogenase, type I | 36362.4 | 517 | 51 | 4.92 | C | GAPDH-I | - 3.34061 | 1.799681 |
| 1476 | 314939418 | - | signal peptide, YSIRK family | 20884.4 | 178 | 31 | 8.69 | CM | - 2.25027 | 0.551905 | |
| 880 | 257882793 | MG | NAD-dependent epimerase/dehydratase | 37782 | 83 | 19 | 5.4 | C | WcaG | - 2.38858 | 0.286398 |
| 569 | 227550654 | F | adenylosuccinate synthase | 47912.6 | 871 | 47 | 5.57 | C | purA | - 1.89042 | 0.132959 |
| 572 | 257900037 | J | tyrosyl-tRNA synthetase | 47550.4 | 288 | 22 | 5.28 | C | - 2.32279 | 0.454633 | |
| 756 | 261207473 | E | phospho-2-dehydro-3-deoxyheptonate aldolase | 37513.2 | 397 | 40 | 5.83 | C | - 2.18216 | 0.784625 | |
| 540 | 69246358 | R | Nitroreductase | 22292.2 | 526 | 72 | 5.05 | C | - 2.30057 | 1.044212 | |
| Protein Up-Regulated in B42-R7 | |||||||||||
| 1356 | 69246358 | R | Nitroreductase | 22292.2 | 526 | 75 | 5.05 | C | 3.495866 | 0.03567 | |
| 566 | 29839250 | G | 2-phospho-D-glycerate hydro-lyase | 46382.4 | 476 | 26 | 4.58 | C | eno | 2.661148 | 0.595582 |
| 692 | 69249876 | G | Glyceraldehyde-3-phosphate dehydrogenase, type I | 36362.4 | 517 | 54 | 4.92 | C | GAPDH-I | 2.242169 | 0.808525 |
| 1609 | 257882505 | - | transcription activator | 17717.9 | 97 | 30 | 4.94 | U | 1.995636 | 0.202806 | |
In addition, we identified the same protein in two spots, which indicated different molecular weights, such as nitroreductase and GAPDH-I. Spot 1356 and spot 540 were identified as nitroreductase. Additionally, GAPDH-I was identified as spot 692 and spot 269. Spot 1356 and 692 were selected as nitroreductase and GAPDH-I by considering pI value, molecular weight and expression level. The identification of two spots as one protein may be due to the existence of homodimers of protein in 2D gels.
Nitroreductase (69246358) was up-regulated in B42-R7. Nitroreductase catalyzes the reduction of nitroaromatic compounds, such as nitrotoluenes and nitrofurans. Studies in E. coli showed that nitrofurans need to be activated by reducing activity for the antibiotic effect. The reduction of activity would thus indicate the nitrofurans sensitivity of the bacteria (3). The induced nitroreductase may explain why B42-R7 became susceptible to nitrofurantoin.
Most metabolism-related proteins that are necessary for growth, especially those related to glycolysis, energy production and conversion, and nucleotide transport and metabolism, were suppressed in the rpoB mutant. For nucleotide transport and metabolism, adenylosuccinate synthetase, which is involved in de novo biosynthesis of AMP, was identified. For synthesis of amino acids, 3-dehydroquinate synthase, which participates in the biosynthesis of amino acids, such as phenylalanine, tyrosine and tryptophan, was analyzed. Phospho-2-dehydro-3-deoxyheptonate aldolase is an intermediate of the synthesis of chorismate from shikimic acid (13). Shikimic acid is a precursor to the aromatic amino acids, phenyl alanine and tyrosine, and was previously obtained by McCalla and Neish (14). Glutathione reductase, also known as GSR or GR, is an enzyme that reduces glutathione disulfide (GSSG) to the sulfhydryl form GSH, an important cellular antioxidant (15). Three of the metabolism-related proteins were induced. GAPDH plays a prominent role in glycolysis in the cytosol. It also participates in tRNA transport, stimulates transcriptional activity, and controls DNA replication and DNA repair (16-19). Additionally, 2-phospho-D-glycerate hydro-lyase participates in glycolysis/gluconeogenesis.
Tyrosyl-tRNA synthetase is involved in protein synthesis. Down-regulated tyrosyl-tRNA synthetase indicated that the bacteria’s capacity to synthesize proteins was limited. The bacteria would logically increase their enzymes such as tRNA synthetase in order to enhance its capacity to synthesize proteins, thus evading the effect of the rpoB mutation. Two proteins without a COG class were identified. They should be investigated further as a potential functional proteins. One of the proteins, an up-regulated transcription activator, and the other protein, a down regulated signal peptide of the YSIRK family.
3.3. Overexpression of Nitroreductase Leads to Sensitivity to Nitrofurantoin
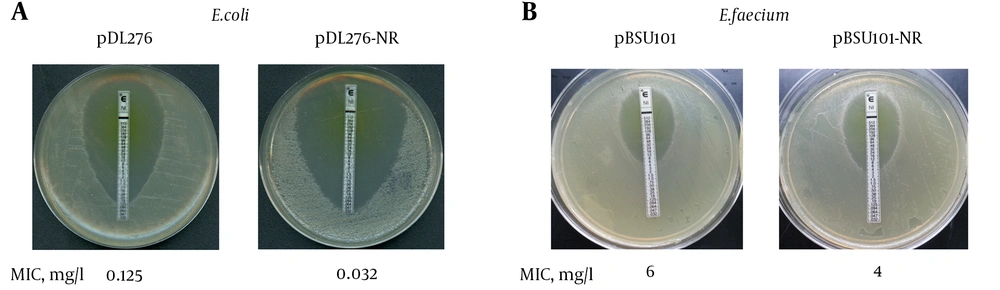
Given that nitroreductase was up-regulated in the rpoB H489D mutant as determined by a 2D-gel, we investigated whether the overexpression of nitroreductase would result in the same phenotype as the rpoB H489D mutant. Recombination expression vectors harboring the nitroreductase gene were introduced to E. coli and E. faecium. The Etest results showed that overexpression of nitroreductase enhanced the sensitivity of both E. coli and E. faecium to nitrofurantoin, indicating that nitroreductase played a role in nitrofurantoin susceptibility (Figure 3).
A, Nitrofurantoin MIC for DH5α harbored pDL276 or pDL276-NR by E-Test; The E. faecium B42 nitroreductase gene was PCR amplified from genomic wild-type DNA and ligated into pDL276 to generate plasmid pDL276-NR; The plasmids pDL276-NR and pDL276 were transformed into E. coli DH5α cells. Nitrofurantoin MICs of DH5α harbored pDL276 or pDL276-NR were determined by Etest on BHI agar; B, Nitrofurantoin MIC for DH5α harbored pBSU101 or pBSU101-NR by E-Test; The nitroreductase gene was sub-cloned into pBSU101 to generate plasmid pBSU101-NR. Both pBSU101 and pBSU101-NR were transformed into E. faecium ATCC 35667; Nitrofurantoin MICs of DH5α harbored pBSU101 or pBSU101-NR were determined by Etest on BHI agar.
4. Discussion
One of the isolated Rifr mutants (rpoB-H489D) of E. faecium presented lower growth rates and higher ROS levels. Therefore, in this study, we used 2-DE to investigate the global proteome altered by the rpoB mutation in E. faecium.
Breeze et al. (20) reported that the precise mode of action of the nitrofurantoin is still not clear. Nitrofurantoin exerts its antibiotic effects through two pathways. First, nitroreductase catalyzes the reduction of nitrofurantoin to form bactericidal end-products, which bind to DNA and proteins and inhibit nucleic acid synthesis. Studies in E. coli showed that nitrofurans required reduction for the antibiotic effect. Sandegren et al. (3) reported that the ability to reduce these compounds indicated the nitrofurans sensitivity of the bacteria. Second, nitrofurantoin had a direct effect on DNA. Breeze et al. (20) reported that DNA repair-defective mutants were also sensitive to nitrofurantoin.
Nitroreductase (69246358) was up-regulated in the rpoB mutant (B42-R7) in proteome analysis. The overexpression of nitroreductase in E. coli provided further evidence that the sensitivity to nitrofurantoin was caused by nitroreductase. Additional effects caused by rpoB mutation, except the rifampicin resistance, have been demonstrated in many bacteria. Ingham et al. (21) reported the rpoB mutations (Q469K and Q469R) not only resulted in rifampicin resistance but also increased sensitivity to regulation by NusG in Bacillus subtilis. Maughan et al. (22) reported that mutations in rpoB of B. subtilis also led to a global change in the expression of a number of global phenotypes known to be under transcriptional control, such as growth, competence for transformation, sporulation, and germination. Gao et al. (23) reported that rpoB H481Y and an active stringent response caused global transcriptional changes and reduced virulence in Staphylococcus aureus. Kristich et al. (24) reported that a H486Y rpoB mutant in enterococci resulted in alteration of intrinsic resistance towards 2nd and 3nd generation cephalosporins. The RNAP mutation affected the transcription of as-yet-unknown gene that was critical for cellular adaptation to cephalosporin stress. The mutant RNAP in E. faecium might mimic the stringent response to regulate the expression of nitroreductase.
The rpoB H489D mutant presented lower growth rates and higher ROS levels. Wells et al. (25) reported that the rpoB mutation restored prototrophy to ppGpp mutants by causing RNAP to mimic its behavior under stringent conditions in Sinorhizobium meliloti. The rpoB mutation mimics the effects of the stringent response by altering transcription at the level of initiation, elongation, or the ability of core RNAP to bind various sigma factors. The lower growth rate provided further evidence that RNAP in the mutant functions as if it were under stringent conditions.
Most of the metabolism-related proteins that are necessary for growth, especially those related to glycolysis, energy production and conversion, and nucleotide transport Most metabolism-related proteins that are necessary in the rpoB mutant. These results were consistent with the lower growth rate. We also found that glutathione reductase (an important cellular antioxidant) was down-regulated (15). The down-regulation of glutathione reductase indicated that the cell may have higher intracellular ROS levels. We verified this hypothesis via ROS measurement in the cells. Hua et al. (7) reported the same result in an rpoB mutant in D. radiodurans. The rpoB mutant in D. radiodurans also showed a lower growth rate and higher ROS levels.
Overall, an rpoB mutant of E. faecium showed a lower growth rate and higher ROS levels. The proteomic profile comparison of E. faecium B42 and its Rifr mutant B42-R7 indicated that the rpoB mutation altered the transcription of nitroreductase and other genes related to growth. The rpoB mutation in E. faecium not only altered the susceptibility to rifampin but also affected global protein expression, including nitroreductase. Thus, nitroreductase in E. faecium B42-R7 might play a role in nitrofurantoin resistance.