1. Background
Recent reports have described that the deoxyribonucleic acid (DNA) virus designated as the SEN virus (SENV) was first discovered in the serum of an intravenous drug user infected with human immunodeficiency virus (HIV) (1). The name for the SEN virus was derived from the initials of the first identified patient (2). SENV has been described as a blood borne infection with a small, circular, single-stranded, non-enveloped DNA virus that contains approximately 3800 nucleotides and belongs to the Circoviridae family, which also includes the Torque teno virus (TTV), TUS01, SANBAN, and YONBAN (3, 4). SENV is sub-grouped into eight distinct strains (A - H) (5, 6). According to previous studies, SENV genotypes H (SENV-H) and D (SENV-D) are the two genotypes suspected of being significantly associated with the pathogenesis of non-A - E hepatitis and also post-transfusion hepatitis (3, 6).
Currently, most investigations have focused on the SENV-H and SENV-D genotypes, which are dominant in different geographical regions of the world (3, 6). Interestingly, SENV is highly prone to mutation, which is a typical feature of RNA viruses but not DNA viruses; these mutations may result in a persistent state of SENV (3, 7). Further studies revealed that SENV characterization is similar to TTV (8). The SENV and TTV genomes have 55% and 37% homogeneity in their nucleotide and amino acid sequences, respectively (2, 3). SENV may be transmitted via a blood transfusion or through mother-to-child transmission (9, 10). The high prevalence of SENV observed among patients with HIV, hepatitis B virus (HBV), and hepatitis C virus (HCV) infections indicate a shared route of transmission (3, 7). As a result, non-transfused individuals are at low risk of contracting an SENV infection (2, 9). Onerecent study reported that the SENV DNA level was not higher in patients with liver diseases than it was in healthy blood donors (11). So far, the pathogenesis of SENV infection remains unknown, and the clinical relevance of SENV alone or in combination with other liver infections is controversial (12).
Occult HBV infection (OBI) is defined as the detection of HBV DNA in the sera or liver biopsy along with a negative result for the hepatitis B surface antigen (HBsAg) with or without anti-HBc or anti-HBs (13, 14). The identification of anti-HBc is important in the diagnosis of 80% of OBIs (15). The emergence of OBI is due to the occurrence of mutations within the α-determinant of the S-gene region of HBV, which is noteworthy because enzyme-linked immunosorbent assay (ELISA) cannot detect the HBsAg in this region (14, 16). As a result, the screening of blood donors using ELISA will miss the OBI; therefore, the gold standard test for OBI detection is a molecular approach, including nested polymerase chain reaction (PCR) or real-time PCR (17). Several studies have suggested that OBI largely occurs in regions with high levels of endemic HBV infection (18). Persistent OBI may result in cirrhosis of the liver or hepatocellular carcinoma (19). In Iran, the incidence of HBV is intermediate, ranging from 1.6% - 3.6% (20, 21). The presence of OBI among blood donors has not yet been reported in Iran. Thus, this study was conducted to determine the incidence of SENV-D and SENV-H infection with a specific focus on OBI diagnosed in blood donors in Ahvaz city, the capital of the Khozestan province, which is located in southwest Iran.
2. Objectives
The aim of this study was to determine the prevalence of SENV-D and SENV-H with a particular focus on OBI diagnosed in blood donors in Ahvaz city, Iran.
3. Patients and Methods
3.1. Serum Samples
This study had a cross-sectional design and included 184 healthy blood donors from the Ahvaz blood transfusion center in Ahvaz city, Iran, from October-November 2013. The sera of all donors were negative for HBsAg, anti-HCV antibodies, and anti-HIV antibodies. All blood samples were also tested for SENV-D and SENV-H DNA using nested PCR. Liver function testing, including aspartate aminotransferase (AST) and alanine transaminase (ALT), and alkaline phosphatase (ALP), HBcIgG (ELISA, DIA.PRO, Italy), and HBV DNA (HbsAg gene) were carried out for each individual donor. This study was approved by the ethics committee of our institution and had the registration number 122 at the Ahvaz Jundishapur University of Medical Sciences. Informed consent was obtained from all participants before the study began. Demographic information was collected from each donor, including age, sex, educational level, living situation, and any history of blood transfusions, surgery, tattooing, cupping, and phlebotomy (Table 1). The following steps were carried out for the nested PCR.
| Variable | SENV-pos variant (%) | SENV-neg | Total No. (%) | P-Valueb | ||
|---|---|---|---|---|---|---|
| D→H→D + H | ||||||
| Age, years | 0.09 | |||||
| < 30 | 1 (3.3) | 4 (13.3) | 0 (0) | 25 (83.3) | 30 (16.3) | |
| 30 - 40 | 4 (5.6) | 11 (15.5) | 0 (0) | 56 (78.9) | 71 (38.6) | |
| 40 - 50 | 2 (3.8) | 9 (17.3) | 0 (0) | 41 (78.8) | 52 (28.3) | |
| > 50 | 3 (9.7) | 8 (25.8) | 2 (6.5) | 18 (58.1) | 31 (16.8) | |
| Level of education | 0.88 | |||||
| Graduate diploma | 9 (6.4) | 24 (17.1) | 2 (1.43) | 105 (75) | 140 (76.1) | |
| AD or BSc | 1 (2.6) | 8 (20.5) | 0 (0) | 30 (76.9) | 39 (21.2) | |
| MSc or PhDd | 0 (0) | 0 (0) | 0 (0) | 5 (100) | 5 (2.7) | |
| Live | 0.27 | |||||
| Ahvaz | 6 (4) | 27 (17.9) | 2 (1.3) | 116 (76.8) | 151 (82.1) | |
| Other | 4 (12.1) | 5 (15.2) | 0 (0) | 24 (72.7) | 33 (17.9) | |
| History of receiving blood transfusions and blood components | 0.17 | |||||
| Yes | 1 (33.3) | 0 (0) | 0 (0) | 2 (66.7) | 3 (1.6) | |
| No | 9 (5) | 32 (17.7) | 2 (1.1) | 138 (76.2) | 181 (98.4) | |
| History of abandonment of addiction (fumigation, eating, or injection) | 0.88 | |||||
| Yes | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 2 (1.1) | |
| No | 10 (5.5) | 32 (17.6) | 2 (1.1) | 138 (75.8) | 182 (98.9) | |
| History of infectious diseases (HBV or HCV) | 0.95 | |||||
| Yes | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (0.5) | |
| No | 10 (5.5) | 32 (17.5) | 2 (1.1) | 139 (76) | 183 (99.5) | |
| History of endoscopic surgery, tattooing, cupping, or phlebotomy | 0.31 | |||||
| Yes | 5 (10.2) | 8 (16.3) | 1 (2) | 35 (71.4) | 49 (26.6) | |
| No | 5 (3.7) | 24 (17.8) | 1 (0.7) | 105 (77.8) | 135 (73.4) | |
aAll blood donors were male. The sera of 8/184 (4.3%) were positive for anti-HBc antibody (2 (4.5%) were SENV-positive, and 6 (4.3%) were SENV-negative), but all were negative for HBV DNA.
bP < 0.05 was considered significant.
cAssociate degree or bachelor degree.
dMasters degree or doctor of philosophy.
3.2. DNA Extraction and Detection of SENV DNA Using Nested PCR
DNA extraction was carried out for all the sera using the high pure viral nucleic acid kit (Roche diagnostics, Germany) according to the manufacturer’s instructions. The extracted DNA was stored at -70°C prior to testing. The forward primer AI-1F (5′-TWCYCMAACGACCAGCTAGACCT-3′) and the reverse primer AI-1R (5′-GTTTGTGGTGAGCAGAACGGA-3′) were used to amplify the target regions of the ORF1 gene of SENV-D and SENV-H by nested PCR (4). For the first round, the 25-µl PCR reaction mixture contained 2.5 μL of a 10X PCR buffer, 0.2 pmol of each primer, 1.5 mM MgCl2, 200 mM dNTPs, 1 U of Smart Taq DNA polymerase (CinnaGen, Iran), and 5 μL of the extracted DNA.
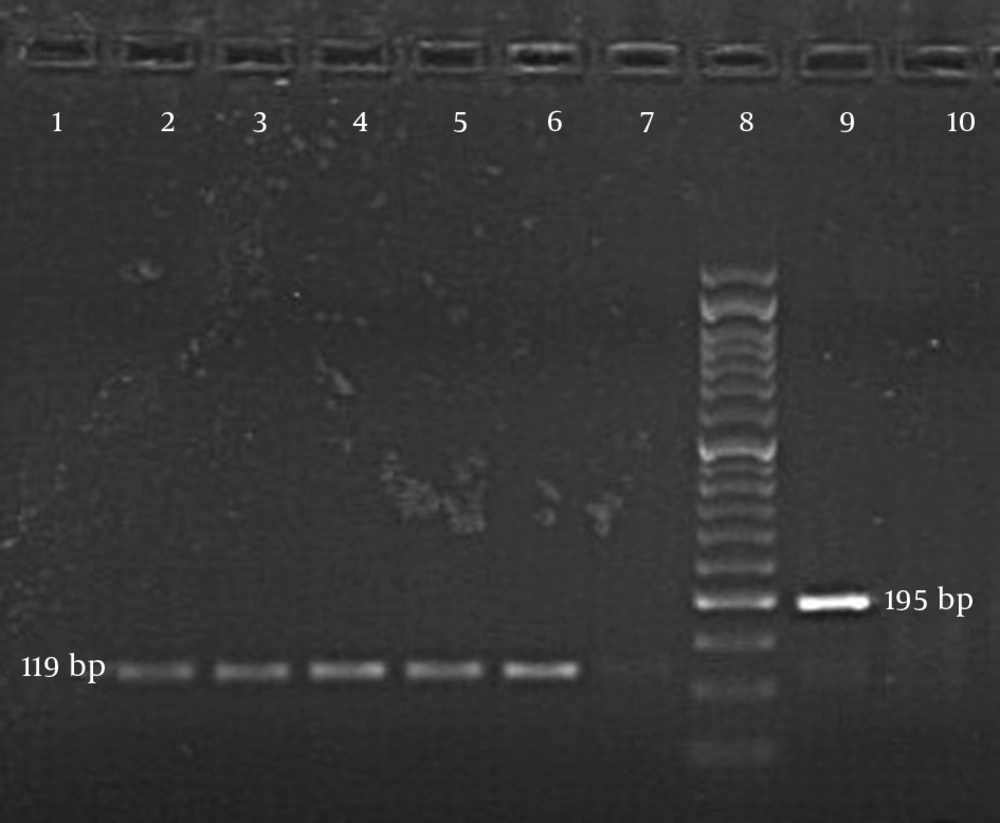
All the samples, including the negative and positive controls, were subjected to a thermocycler (Peqlab, Germany) and programmed with the following conditions: 94°C for 5 minutes and 35 cycles, 94°C for 1 minute, 53°C for 1 minute, and 72°C for 1 minute with a final extension at 72°C for 10 minutes. For the second PCR round, 3 μL of the product from the first PCR round were used. The D-1148F (5′- CTAAGCAGCCCTAACACTCATCCAG-3′) and D-1341R (5′-GCAGTTGACCGCAAAGTTACAAGAG-3′) primers were used for the SENV-D genotype. For the SENV-H genotype, forward primer H-1020F (5′-TTTGGCTGCACCTTCTGGTT-3′) and reverse primer H-1138R (5′-AGAAATGATGGGTGAGTGTTAGGG-3′) were used (4). The contents of the second PCR reaction mixture were similar to those used in the first round. The PCR conditions were programmed at 94°C for 5 minutes, 25 cycles of 94°C for 1 minute, 54°C for 1 minute, and 72°C for 1 minute. A final PCR product of 195 base pairs indicated a positive reactivity for SENV-D, while 119 base pairs revealed the same for SENV-H after appearing on 1.5% agarose gel electrophoresis supplemented with a DNA-safe stain (CinnaGen, Iran).
3.3. Nested PCR for HBV DNA
Nested PCR was used to detect HBV DNA in the sera of all blood donors. For the first round, forward primer SA1: (5′-ATCGCTGGATGTGTCTGCGG-3′) and reverse primer SA2 (5′-GGCAACGGGGTAAAGGT TCA-3′) were used (22). The 25-ul PCR reaction mixture contained 7 μL of the template, 2.5 μL of the 10x PCR reaction buffer, 1 μL dNTP (10 mM), 0.75 μL MgCl2 (50 mM), 0.5 μL forward/reverse primer, 0.2 Taq DNA polymerase (CinnaGen, Iran), and 12.5 μL water. The negative, positive, and control samples were subjected to a thermocycler (Peqlab, Germany) at 94°C for 5 minutes and 35 cycles, 94°C for 1 minute, 54°C for 1 minute, 72°C for 1.5 minute, and a final extension at 72°C for 10 minutes. For the second round, 5 μL of the first-round product was used for the template, and the inner forward primer SB1 (5′-TTAGGGTTTAAATGTATACCC-3′) and inner reverse primer SB2 (5′-CATCTTCTTGTTGGTTCTTCTG-3′) were used (22). The PCR reaction mixture contents were similar to those used in the first round, and the following PCR conditions were programmed: 94°C for 5 minutes, 30 cycles at 94°C for 1 minute, 45°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 10 minutes. The PCR products were analyzed on 1.5% agarose gel electrophoresis stained with a DNA-safe stain (CinnaGen, Iran) and photographed under ultraviolet light. The presence of a PCR product with 416 base pairs indicated a positive reaction.
3.4. DNA Sequencing
Ten positive PCR products of SENV were randomly selected, purified, and sent for sequencing by 1st BASE laboratories in Malaysia.
4. Results
All blood donors were male, and their ages ranged from 30 - 62 years, with a mean age of 40 ± 9.4 years. AST, ALT, and ALP results among the healthy blood donors were within the normal range. The results of the nested PCR following agarose gel electrophoresis indicated a 195-base pair band for SENV-D and a 119-base pair band for SENV-H (Figure 1). The incidence rates of SENV-D and SEV-H among the blood donors were 10/184 (5.4%) (95% confidence interval (CI): 2.1% - 9%) and 32/184 (17.4%) (95% CI: 12% - 23%), respectively. There were 2 (1.1%) coinfection cases of both SENV-H and SENV-D among the blood donors. SENV viremia was identified in 44 (24%) of the 184 blood donors (95% CI: 18% - 30%). The prevalence of SENV-H was 3.2-fold higher than that of SENV-D in healthy blood donors. The incidence of anti-HBc antibody was 8/184 among the blood donors (4.3%).
All the blood donors’ sera, including those positive for the anti-HBc antibody, were negative for HBV DNA. Thus, OBI was not detected among the blood donors. The results of homology sequencing of SENV-D and SENV-H in the 10 samples varied from 91% - 100%. The sequence homology was 91% among the isolated SENV-D cases (GenBank: accession numbers AB856069.1 and GQ179970.1), while the sequence homology was 100% among the isolated SENV-H patients (GenBank: accession numbers GQ452051.1 and GQ452053.1). The prevalence of SENV and variables such as age, education, location, and history of blood transfusion and surgical endoscopy, were not significant (P > 0.05) (Table 1).
5. Discussion
In our study, SENV-D and SENV-H had an incidence of 5.4% and 17.4%, respectively, among healthy blood donors. Two (1.1%) blood donors had co-infection with SENV-D and SENV-H variants. Overall, 44 (24%) of the blood donors were positive for both SENV-D and SENV-H viruses. Our results are consistent with the findings of 24% SENV-D and SENV-H reported in Greece (23) and also the results of 23.08% that were reported by Sharifi et al. in Tehran in 2008 (24). The prevalence of SENV strains D and H among healthy blood donors varied from a low of 1.5% in the United States (7) to a high of 90.8% in Iran (25). The incidence of SENV has also been reported in China (31%) (26), Japan (10% - 22%) (10, 27), Germany (17%) (28), Taiwan (15%) (29), Italy (13%) (30), and Thailand (5%) (31). Out of the eight possible SENV variants, most studies have focused on the SENV-H and SENV-D variants because they are the only two blood borne viruses (7, 10, 26). In our study, only 1 (2.3%) of the 44 donors who were positive for SENV had a history of blood transfusion; the remaining 43 (97.7%) did not. The prevalence of SENV-H was also 3.2-fold higher than that of SENV-D in healthy blood donors. In some recent studies, the prevalence of SENV-H was 10 times higher than the SENV-D variant in patients with chronic hepatitis B or C and also in patients with non-A - E hepatitis (7, 8, 29).
In our study, regardless of the incidence of 24% SENV cases, no liver function test abnormalities (AST and ALT) and or abnormal ALP levels were observed among the healthy blood donors. However, no pathogenesis of SENV in the liver biopsies has been reported. Co-infection with SENV and other viral hepatitis infections (HBV and HCV) has been observed in high-risk group patients and also in healthy blood donors (3, 25, 26). In 2010, Karimi-Rastehkenari et al. reported that the frequency of SENV-H or SENV-D and co-infections of HBV or HCV were significantly higher among thalassemic patients (98%) than in healthy individuals (90.8%) in Iran (25). Kao et al. concluded that the rate of SENV-D and SENV-H in thalassemic patients (90%) was greater than that of parenteral drug users (54%), hemophilic patients (68%), and hemodialysis patients (68%) (29).
Therefore, the outcomes of these aforementioned studies indicate that SENV-D and SENV-H are blood borne infections (7, 26, 29). Omar et al. reported the prevalence of SEN virus infection in Egypt in patients with chronic HCV (13.5%) and also in hemodialysis patients (11.1%); similar results were found in the control group (9). Therefore, they suggested that another route of SENV transmission should be further investigated (24, 28). Like TTV (32), SENV may be transmitted via a commensal relationship among hosts with no obvious clinical features (24).
Recently, due to the occurrence of mutations in the HBsAg gene, a new type of HBV infection named OBI has emerged. One of the main transmission routes of OBI is through blood transfusions or blood products (33, 34). OBI is defined as the detection of HBV DNA in the serum or liver tissue along with undetectable HBsAg; these patients may also test positive for HBcIgG (35). Occult HBV has spread worldwide and is largely prevalent in endemic regions (36). The study of Kafi-abad et al. in Tehran (2004 - 2007) reported that no OBI cases were found among the blood donors (37). In contrast, the prevalence of OBI among blood donors in Mexico, Taiwan, and China was reported to be 6.4%, 4%, and 9.8%, respectively (38-40).
In conclusion, the results of this study indicated that 24% of healthy blood donors have been infected with SENV-H or SENV-D. Although 8 (4.3%) of the healthy donors tested positive for HBcIgG and negative for HBV DNA, a liver biopsy for HBV DNA detection is recommended to confirm the final OBI diagnosis.