1. Background
Candida species are the predominant cause of fungal infections worldwide. They cause non-life-threatening mucocutaneous illnesses as well as invasive infections that can virtually affect any organ. This species of yeast mainly exists as harmless commensal flora on the human skin and in the gut. However, when the immune system is suppressed, they may become pathogenic and cause chronic mucocutaneous and invasive infections. Such events are common in intensive care units and among neutropenic patients (1-4), and despite the substantial advancements in the prevention strategies of hospital-acquired infections, fungi belonging to the genus Candida have remained the fourth most commonly isolated nocosomial blood pathogens, associated with a fatality rate of 5% - 71%. This is a very alarming rate considering that between the years 2000 and 2005, the incidence of Candida albicans was found to have almost doubled. Infections with other species, notably C. glabrata, C. parapillosis, C. krusei, and C. tropicalis, were also found to have generally increased in the recent years (1, 5-8).
The pathogenicity of C. albicans is predominantly because of specific genes enabling the fungal cells to colonize the host and cause diseases affecting the skin, gastrointestinal tract, and oral cavity. Biofilm formation is an important virulence factor of C. albicans. A biofilm is a heterogeneous structure composed of hyphae, pseudohyphae, and yeast cells, and it develops at the interface between an aqueous medium and a solid. In the past two decades, the increased use of medical implant devices has led to an increase in the rate of Candida infections (9-11). Experimental studies revealed that biofilm formations on these devices allowed fungal cells to encapsulate and aggregate, thus forming phenotypically and morphologically distinct microcolonies that were extremely tolerant of antimicrobial agents in comparison to free fungal cells. The aggregated cells were often enclosed in a self-produced extracellular matrix (ECM) that conferred protection from the immune system of the host and antimicrobial agents a mechanism that substantially increased the survival rate of the cells (5, 9, 12-14).
2. Objectives
The present study is a comprehensive review of literature conducted to identify the resistance mechanisms of C. albicans biofilms to drugs and the host immune system.
3. Search Strategy
Fungal biofilms have been implicated in the pathogenesis of a large number of invasive, disseminated infectious diseases. Recent studies have provided insight into the complex processes of biofilm formation. Candida albicans has been one of the most studied fungi. Many different in vitro models of the organism on various potential growth surfaces, including medical devices, have been used to study the structure and architecture of biofilms to understand their mechanisms of resistance towards antifungal agents (15-17).
4. Biofilm Development
Biofilm formation is found to occur in a variety of systems. According to Costerton et al., in all self-sustaining aquatic ecosystems, bacteria predominantly grow in biofilms, and cells found in such structures significantly differ from their planktonic counterparts (cells in suspensions) (18).
A biofilm is a dynamic environment consisting of microcolonies of cells enclosed within a highly hydrated self-produced ECM. Research seems to indicate that biofilm formation is a defense mechanism against nutritional and environmental stress. It was shown to involve the expression of a large number of transcriptional factors and non-/specific genes (12, 19).
A key characteristic of biofilms is their resistance to broad-spectrum antimicrobial agents. For example, the comparison between yeast cells in their planktonic state and cells in biofilms showed that biofilm formation resulted in resistance to several antifungal agents, including azoles and echinocandins. Biofilms were found to be tolerant of amphotericin B and fluconazole, two of the most commonly used systemic antifungal drugs for the treatment and prophylaxis of systemic candidiasis. In particular, decreased biofilm susceptibility to fluconazole in C. glabrata and C. krusei isolates resulted in a significant increase in the patient mortality rate (20).
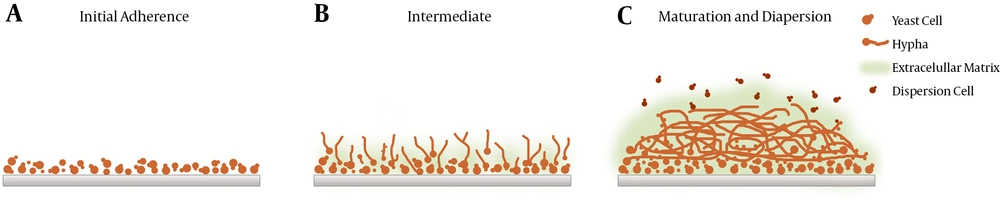
To reach maturity, biofilms undergo three developmental phases (early, intermediate, and mature) involving complex and successive growth processes marked by increased cellular metabolic activity (5, 13) (Figures 1 and 2). Mature biofilms consist of dense networks of cells in the form of yeasts, hyphae, and pseudohyphae joined together by a polymeric ECM. Water channels between the cells facilitate the diffusion of nutrients from the environment to the lower layers of the biomass and allow wastes to be efficiently eliminated.
As yeasts are rapidly becoming resistant to available medications, the identification of key factors that contribute to biofilm complexity and resistance has become vital for successful treatment (15). However, thus far, the factors responsible for the resistance of Candida biofilms to antifungal agents have been elusive. Although collective characteristics and synergistic interactions between species have known to result in resistance in most cases, C. albicans resistance may rather be due to the following factors: i, The complex architecture of biofilms and their associated ECM, which significantly hinders drug diffusion; ii, increased metabolic activity and increased expression of drug efflux pumps (channels that expel harmful compounds from the colony); and iii, metabolic plasticity, which allows sessile cells to undergo metabolic changes in response to environmental and nutritional stress (5, 21-23).
Candida albicans biofilms are mainly bi-layered. The bottom layer commonly comprises yeasts tightly attached to the surface, whereas the upper layer is usually made up of hyphae. Nonetheless, the final architecture of biofilms has shown to vary according to the substrate used for energy and the growing conditions (24). Biofilms evolve similarly, whether in vivo or in vitro. However, in vivo models of C. albicans biofilms have shown to mature more rapidly and develop substantially thicker walls.
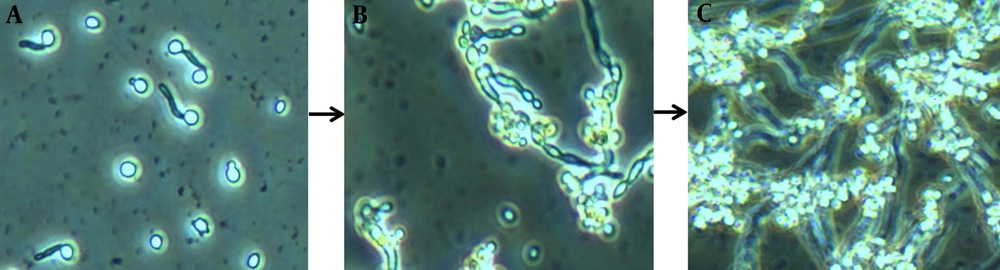
These images depict biofilm formation on a polystyrene surface under static conditions (25). A, Biofilm formation is initiated as yeast cells attach to a potential growth material and develop a consistent biomass; B, greater cellular density results in increased metabolic activity as the period of cultivation is extended; C, clusters of heterogeneous microcolonies are reinforced by the production and accumulation of a slime-like extracellular matrix (ECM). Maturity is reached as ECM encloses the sessile yeast and filamentous cells.
Candida albicans Biofilms at Three Different Developmental Phases: Early, Intermediate, and Mature (25)
5. Biofilm Complex Architecture
Candida biofilms are extremely tolerant towards common antifungal agents and the immune system. The biofilms’ enhanced resistance to drugs makes the surgical replacement of infected devices (venous catheters, urinary catheters, pacemakers, and artificial joints) the only option (26). Considering that over ten million patients receive such devices annually, noninvasive therapeutic strategies are highly sought after (9, 27).
Candida albicans biofilm formation starts with free yeast cells being attached to an abiotic surface, such as an implanted medical device. Owing to their high surface hydrophobicity, C. albicans cells are capable of virtually attaching to any surface. Attachment is accompanied by the upregulation of the genes responsible for cell wall formation, e.g., HWP1 and ALS1 genes (5). This is followed by proliferation characterized by mixed growths of unicellular yeast cells (pseudohyphae). This phase is detected by the visualization of hyphae across the biofilm surface. According to Nett et al. (28), as the biofilm matures, cells undergo morphological transition to create a mesh consisting of ECM and different growth forms. The accumulation of ECM in mature cells results in resistance to antimicrobial agents and the host’s immune system (9) as ECM shields the enclosed cells from attacks (29). Research indicated that the main component of ECM in C. albicans biofilms was β-1, 3-glucan (30).
6. Metabolic Activity
Morphological and phenotypic differences between sessile and free cells are associated with different levels of gene expression. For instance, glucan synthase gene (FKS1) is found to be upregulated during biofilm formation. Multidrug resistant phenotypes of C. albicans were shown to have associated with elevated cellular levels of proteins encoded by CDR1, CDR2, and MDR1 receptor genes (31, 32). The overexpression of CDRI and CDR2, multidrug transporters of the ABC family, is linked to diminished yeast susceptibility to azole antifungal agents. The expression of MDR1, a P-glycoprotein responsible for the ATP-dependent expulsion of certain compounds, has also been found to be high in drug-resistant biofilms. However, according to Ramage et al. (33), efflux pumps do not contribute to biofilm antifungal resistance. Although efflux pump genes expression was vital to the formation of biofilms, research showed that expression levels were not altered with the use of antifungal agents and did not confer the resistance of sessile cells. Observing the effects of mutant and wild-type efflux pump genes on the biofilm resistance to drugs, Mukherjee et al. (32) reported that although the structure of the biofilms containing the mutant genes was similar to the wild-type biofilms, the former was susceptible to antifungal agents at the early phases of formation. Resistance was rather found to develop later as the biofilms matured. The wild-type biofilms, however, exhibited reduced susceptibility to antifungal agents in all formation phases. This suggested that efflux pumps conferred resistance only in the early phase of formation. Bruzual et al. (34) held that upregulated expression of efflux pumps could not account for the resistance of biofilms in the presence of high concentrations of antifungal drugs. Watamoto et al. (31) believed that even though the expression of efflux pump genes was not essential for resistance, it facilitated the acquisition of resistance.
The mature biofilm ECM is primarily composed of β-1, 3-glucan. Other components such as proteins and phosphorous were found to strengthen cell adhesion and protect sessile cells from antifungal agents through known and unknown mechanisms. The synthesis of β-1, 3-glucan is dependent on the expression levels of glucan synthase (FKS1). FKS1 acts by triggering certain transcriptional factors to deliver glucan to the matrix in a controlled manner. The downregulation of this gene has been associated with reduced glucan levels and lower activity of glucan-modification enzymes. Therefore, the upregulation of FKS1 may serve as a protective mechanism against stress caused by antifungal drugs (9, 35). However, according to Taff et al., effective matrix glucan delivery and arrangement depend not only on FKS1 but also on the effects of glucan transferases and exogluconases (9). Compared with the wild type, single, double, and triple knockout glucan mutant strains (Bgl2-/-, xog1-/-, and phr1-/-, respectively) were found to have significantly lower matrix glucan concentrations and showed higher sensitivity to triazole. On the other hand, investigating the effects of kinase C pathway regulators on biofilms formed by Saccharomyces cerevisiae and C. albicans, Nett et al. (28) noted that albeit having no impact on the development of a resistant phenotype, the activation of homologs of kinase C regulators induced FKS1 and β-1, 3-glucan synthesis.
7. Metabolic Plasticity
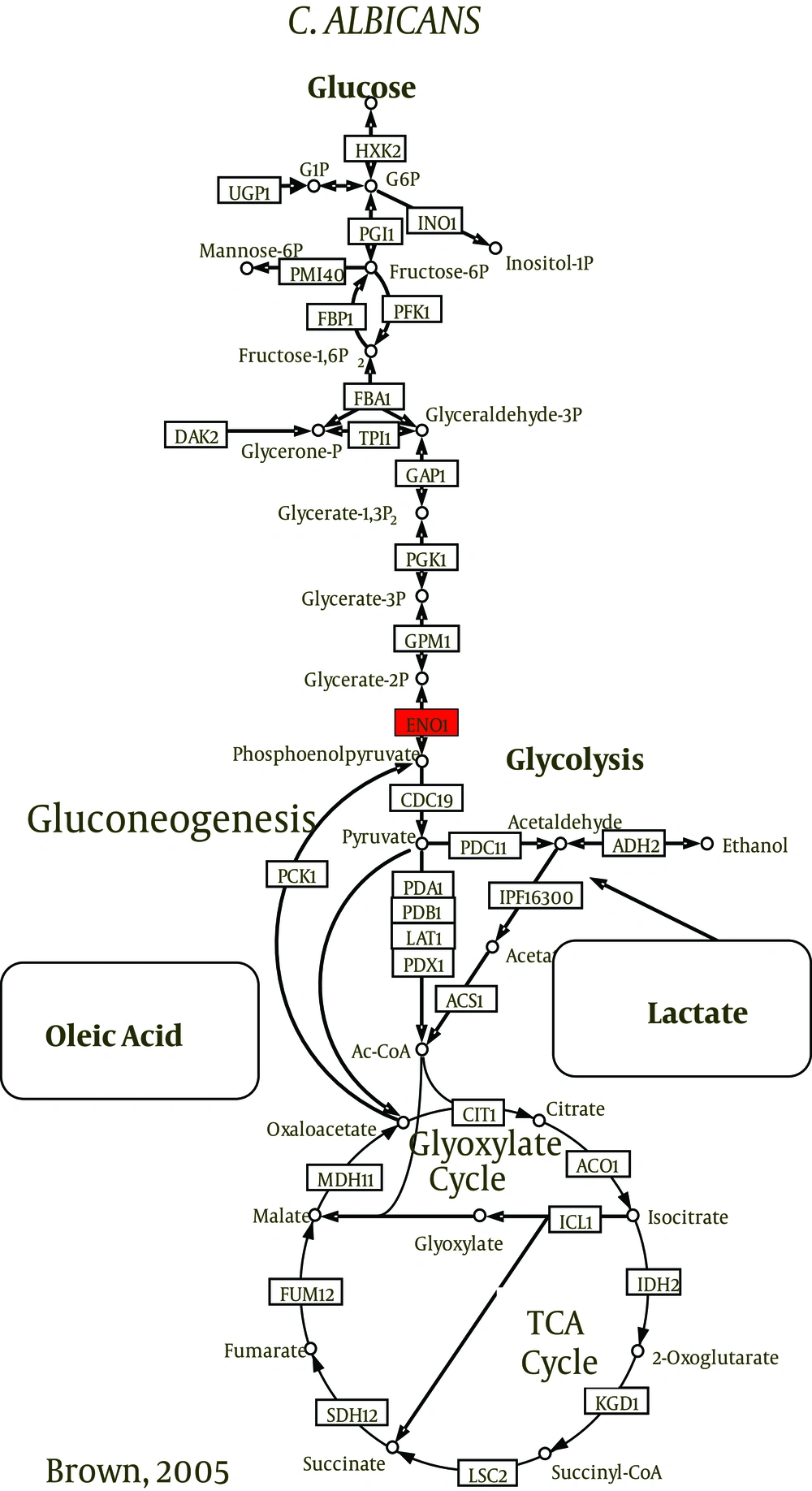
Carbon is vital to the survival and growth of all organisms. To survive within the host, C. albicans should be able to utilize glucose, lipids, proteins, amino acids, and other alternative carbon sources and rapidly adapt to changes in nutrient availability (22). Studies have shown that C. albicans could switch back and forth between glycolysis and gluconeogenesis depending on the conditions of the medium. To generate energy when found in the blood stream, the fungal cells use glucose via glycolysis and TCA cycles. Cells engulfed by phagocytes, however, resort to gluconeogenesis in response to glucose-depletion (36). They also activate the glyoxylate cycle that allows them to utilize the proteins, amino acids, lipids, and phospholipids available inside phagocytic cells (37). These properties make C. albicans quite resilient. Previous reports have shown this metabolic plasticity of C. albicans to be crucial for survival and growth and as a determinant of the organism’s pathogenicity (22, 38, 39).
The utilization of carbon sources is fundamentally important for the species to be infectious in human hosts (40). Candida albicans was found to alter the expression of its metabolic functions to promote cell survival (41) and upregulate amino acid biosynthesis genes to grow efficiently in biofilms (42). Moreover, when exposed to human neutrophils or cultured macrophages, the fungal cells were shown to upregulate amino acid biosynthesis genes and switch from fermentative to nonfermentative metabolism (37, 41, 43). The utilization of nonfermentable carbon sources is achieved by the activation of gluconeogenesis and the glyoxylate cycle (44) that is considered a perquisite for virulence (45). The pathogenicity of C. albicans is dependent on metabolic flexibility and adaptation (38).
The central carbon metabolism pathways such as glycolysis, gluconeogenesis, pentose phosphate pathway, tricarboxylic acid (TCA) cycle, and glyoxylate cycle are highly conserved (46, 47). Candida albicans is labeled as a Crabtree-negative yeast (48) as it retains its respiratory capacity when excess glucose is present (49). It was shown to be sensitive even to very low levels of glucose (50, 51). Glycolytic genes were upregulated and gluconeogenic and TCA cycle genes were downregulated in response to only 0.01% of glucose.
8. Results
The metabolism of Candida species is subject to environmental signals. Glucose is the preferred source of energy for the fungi and is utilized by glycolytic enzymes. Glycolysis is an essential mechanism for morphogenesis and virulence. However, to survive in stressful conditions, fungal cells tend to utilize nonfermentable carbon sources. The consumption of these sources is attempted through highly conserved catabolic and anabolic biochemical reactions that release energy and diffusible quorum sensing molecules (QSMs). Interestingly, these molecules contribute to some extent to the overall resistance of fungal biofilms as the attenuation of carbon metabolism has shown to suppress the virulence of free cells (22, 37, 52, 53). Depending on whether growth occurs on nutrient-laden surface or a nutrient-depleted surface, it has been found that different surface genes have activated in biofilms (12).
Candida species utilize alternative pathways (glyoxylate and gluconeogenesis cycles) to oxidize nonfermentable carbon sources. Recently, it has been found that a couple of enzymes were involved in the process: isocitrate lyase enzyme (ICL1) and phosphoenolpyruvate carboxylkinase enzyme (PCK1) (54, 55). The investigations of energy production mechanisms of biofilms and planktonic cells grown in nutrient-depleted media (stressful environment) reveal that glyoxylate- and gluconeogenesis-related transcripts of ICL1 and PCK1 were markedly more abundant in mature biofilms (23, 30, 56, 57). According to Nobile et al. (58), alternative metabolic pathways resulted in hexose byproducts. These monomers may indirectly contribute to the overall resistance of biofilms because they represent the building blocks for the synthesis of β-1, 3 glucan.
Biofilms were found to be resistant to antifungal therapy primarily because of their glucan-based ECM and impermeable structures. However, recently, C. albicans resistance and candidiasis recurrence were found to be strongly associated with a particular group of cells called persister cells (59). These cells were found to be metabolically dormant in biofilms, but highly resistant to antifungal agents (60). They remain unaffected as the antifungal drugs and the host immune system attempt to eliminate C. albicans biofilms, and alarmingly, after the discontinuation of antifungal agents, research showed that biofilms were repopulated by these highly resistant cells (61, 62).
9. Conclusion
Invasive Candida infections are due to the species’ ability to form biofilms. The treatment of such infections is not straightforward as long-term antifungal therapy may result in the mutation of susceptible genes and increased prevalence of drug-resistant Candida. Furthermore, the accumulation of quorum-sensing molecules may trigger certain genes and produce biofilm resistant phenotypes. Further studies elucidating the correlations between metabolism, resistance, and virulence may eventually provide the potential therapeutic mechanism to suppress drug-related mutations. Moreover, studies on interorganism biofilm formation and antimicrobial resistance may provide further insight into cooperative cohabitation and the behavior of resistant biofilms.