1. Background
Free-living amoebae (FLA), from the genera of Acanthamoeba, Balamuthia and Naegleria can be found all around the world (1). Acanthamoeba spp., are opportunistic protozoan parasites that can be isolated from the wide variety of environments such as tap, fresh, coastal, bottled mineral waters, contact lens solutions, eyewash stations, soil, dust, sewage, ventilators and air-conditioning units (1-4). Some species of Acanthamoeba have direct roles as opportunistic human pathogens causing amoebic keratitis, especially in contact lens wearers and life-threatening granulomatous amoebic encephalitis (GAE) in immunosuppressed/compromised patients, such as HIV/AIDS and organ transplant recipients (5). It is shown that more than 80% of the normal human population has antibodies against Acanthamoeba in their body. This is clearly indicative of that the organism often comes into contact with humans (6, 7).
Acanthamoeba has two forms in its life cycle, active trophozoites and resistant cysts. The double-layered coats of cysts enable resistance against disinfectants, such as chlorine compounds and antibiotics. Moreover, it can survive in a wide range of temperature variations, -2°C to 45°C (5, 8). Based on the 18S rRNA gene sequences, the genus Acanthamoeba is classified into 17 different genotypes (T1 - T17). The majority (90%) of infection-causing isolates belong to the T4 genotype (9, 10) and more than ninety percent (> 90%) of the Acanthamoeba isolates that produce keratitis belong to the T3 and T4 genotypes (11, 12). Of the genotypes (T4, T3, and T2) associated with Acanthamoeba keratitis (AK), T2 and T4 have been reported in Iran’s water (10, 13).
The different species of Acanthamoeba are classified based on the size of cysts and the numbers of arms into the three distinct morphological groups (I, II, and III); group I: with an average diameter of cysts ≥ 18 μm, fewer than six arms and smooth ectocyst and star shape endocysts, group II: it is possible for the ectocyst and endocyst to be close together or widely separated, ectocysts may be thick or thin and endocysts may be polygonal, triangular or round with a mean diameter of less than 18 μm, group III: ectocysts are thin and endocysts may have 3 - 5 blunt corners with the mean cyst diameter of < 18 μm (9, 14, 15). In recent years, remarkable focus has been dedicated to Acanthamoeba due to (i) increasing Acanthamoeba infections, (ii) the potential role of Acanthamoeba in the ecosystem, (iii) model organism to study the molecular biology of motility and phagocytosis resemblance to macrophages and (iv) being a host or reservoir for microbial pathogens (15).
2. Objectives
Although several studies have been accomplished on molecular characterization of Acanthamoeba spp. in different regions of Iran (16, 17) there is no similar research that has been done in the our study area, therefore, information on the occurrence and distribution of Acanthamoeba spp. in environmental and drinking water in West Azerbaijan province remains unclear. The aim of the present study was to determine the epidemiological status of Acanthamoeba spp. based on their molecular and morphometric features in water sources of the West Azerbaijan province, northwest Iran.
3. Methods
3.1. Study Area
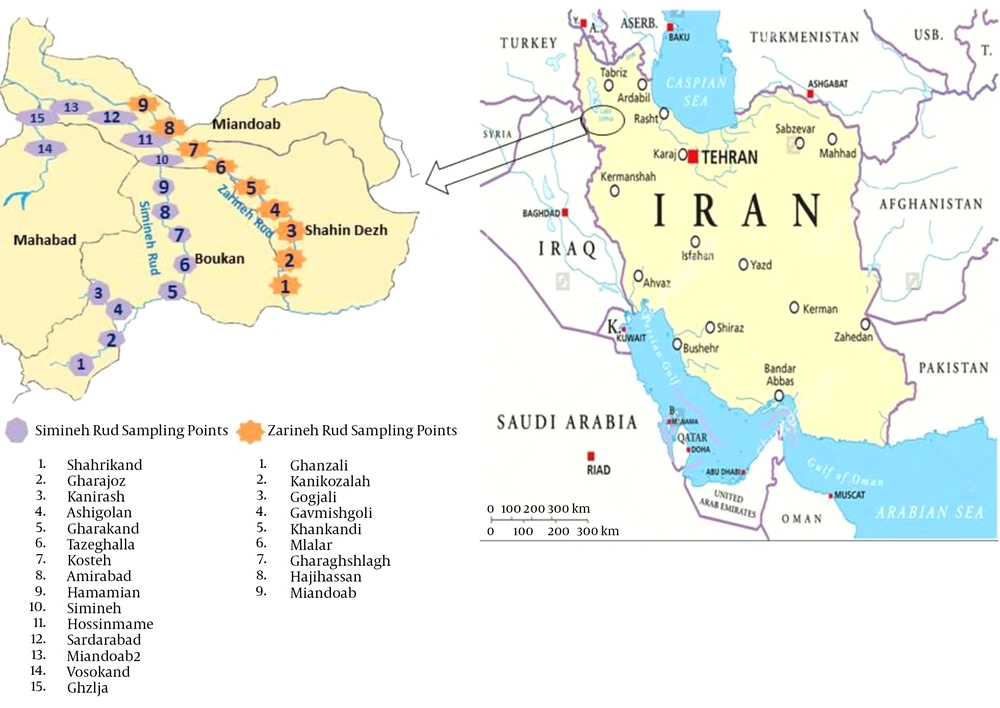
In this cross-sectional study, 60 water samples were collected during June to September 2015 from river sources and tap water in the southern parts of the West Azerbaijan province. The water samples were collected from different parts of Simineh Rud (28 samples) and Zarineh Rud (16 samples), the two main rivers located in the West Azerbaijan province, as well as from the tap waters of the cities of Boukan (four samples), Shahindezh (four samples), Miandoab (four samples) and Mahabad (four samples) (Figure 1).
At the time of sampling, free residual chlorine was measured by the Cl D.P.D Amcor test Kit (HACH, Germany). Each sample (1000 mL) was filled in a sterile glass bottle and transferred immediately to the Parasitology Department of Tabriz University of Medical Sciences, Iran, within 24 hours, in room temperature and stored at the same temperature for subsequent analysis.
3.2. Acanthamoeba spp. Isolation and Measurement
All volume of each water sample (1 liter) was passed through a cellulose nitrate filter, 0.42 µm in diameter (Millipore, Germany) with a weak vacuum (flow rate, 1.3 mL/minute). The samples were divided into two parts and filtered separately using a filtration system. One of the filters was placed directly on the culture medium (1.5% non-nutrient agar plates) and the other was shaken in Page’s Amoeba Saline and then the resulting solution was centrifuged for 10 minutes at 500 rpm. The sediments were cultured on 1.5% non-nutrient agar (Merck, Germany) plates seeded with heat-killed Escherichia coli and then incubated at 37°C for up to 14 days. During the incubation period, the amoebic growth was examined by inverted microscope (Olympus, Japan) on a daily basis. The positive amoebic growth cultures were examined based on morphological characteristics for the presence of trophozoites and cysts, particularly considering their two-layered cell wall cysts.
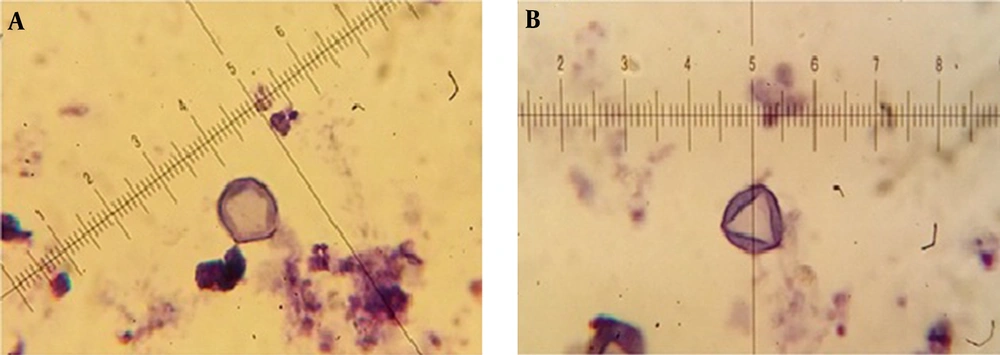
Isolated Acanthamoeba spp. were transferred into axenic culture, peptone yeast extract glucose broth (PYG) (Sigma-Aldrich, USA) [0.75% proteose peptone (wt/vol), 0.75% yeast extract (wt/vol), and 1.5% glucose (wt/vol)]. The identification of Acanthamoeba at the genus level was performed according to the distinctive features of double-walled cysts. Microscopic slides were prepared from positive cultures and stained by Giemsa’s staining method as followed: the fixed smear was immersed in Giemsa (Merck, Germany) solution (1: 45 mL of dH2O) in a staining container for 60 minutes. The slide was then rinsed with dH2O (dried at 37°C for 60 minutes) and studied under light microscope (BH-2, Olympus Corporation, Japan) using 100X objective with scale bar. The diameter of the cysts and the numbers of arms were recorded and classified based on the Pussard and Pons classification (18). Different characters of cysts were measured by ocular micrometer, which were previously calculated by dividing ocular micrometer to stage micrometer (100 × objective = 1 µm per unit space).
3.3. DNA Extraction
DNA extraction kit (Takapou Zist, Iran) was used to extract the total genomic DNA in the concentrated debris according to the kit protocol. The purity of extracted DNA from each sample was determined by the NanoDrop (ND-1000, Termo Fisher Scientific, USA) in the wavelength of 260 nm.
3.4. Polymerase Chain Reaction
A pair of primers (JDP1–JDP2); JDP1: 5’-GGCCCAGATCGTTTACCGTGAA-3’ and JDP2: 5’-TCTCACAAGCTGCTAGGGGAGTCA-3’ was used to amplify a fragment of approximately 500 bp Acanthamoeba genus-specific 18S ribosomal DNA (19, 20). PCR was performed in a 30 μL volume reaction mixer containing 1.25 U of Taq DNA polymerase (Cinnagen, Iran), 2 μL of template DNA and 0.3 mM dNTP, 20 pmol of each primer, 1.5 mM MgCl2 and 10 mM PCR buffer. PCR was performed using an initial denaturation condition for 3 minutes at 94°C, 32 amplification cycles (denaturing 94°C for 45 s, annealing at 55°C for 35 s, and extension at 72°C for 1 minute) and a final elongation step for 10 minutes at 72°C. The PCR products were fractionated by gel electrophoresis on 2% agarose gel that the DNA Safe stain (Invitrogen, UK) has added to the gel solution during preparation and the ampliqons were visualized under UV light in a transilluminator device.
3.5. 18S rRNA Sequencing and Phylogenetic Analysis
In this study, in order to get the genotyping and phylogenetic analysis, the Genetic Analyzer 3130 ABI sequenced 14 isolates of Acanthamoeba. Ambiguous (heterozygous) sites were coded using the standard IUPAC codes for combinations of two or more bases. Contigs from all samples were aligned and edited in consensus positions compared to GenBank sequences of all regional species using the Sequencher Tmv.4.1.4 software for PC (Gene Codes Corporation). The MEGA 5.05 software and maximum likelihood algorithm with kimura 2-parameter model for phylogenetic analysis were used. The accuracy of phylogenetic tree was evaluated by 1000 bootstrap re-sampling.
4. Results
The mean concentration of free residual chlorine was about 0.3 - 1 mg/Liter. Based on the morphological identification, 27 (45%) out of 60 collected water samples were positive to Acanthamoeba spp. using both culture and morphological examinations. Based on Pussard and Pon’s criteria (18), 19 cases belonged to group II (70.4%) and 8 cases were members of group III (29.6%). It should be mentioned that co-infections with both groups was identified in one sample. The ranges of identified cysts diameters were measured 11.1 ± 1.02 to 16.4 ± 4.76 μm (Table 1). Giemsa staining method was used for morphological study (Figure 2). All of the positive samples were from the rivers and the amoeba was not observed in the any of the tap water samples. PCR was performed using JDP1 and JDP2 primers for small subunit ribosomal DNA and detected approximately 500 bp band on agarose gel for all positive isolates. Fifteen of the positive PCR samples (53.5%) were taken from Simineh Rud river and nine samples (56.5%) were taken from Zarineh Rud river. There was no positive sample in the studied tap water samples (Table 2).
| No. | Region | Cyst Diameter, μm | Groupb | Culture | PCR | Genotype |
|---|---|---|---|---|---|---|
| 1 | Gharakandc,d | 11.1 ± 1.02 | II | + | + | T4 |
| 2 | Amirabadd | 14.2 ± 0.82 | II | + | + | - |
| 3 | Hamamiand | 11.1 ± 1.2 | III | + | + | - |
| 4 | Hossinmamed | 12.1 ± 0.99 | II | + | + | - |
| 5 | Miandoabe | 11.8 ± 1.01 | III | + | + | - |
| 6 | Kostehd | 11.3 ± 1.01 | III | + | + | T4 |
| 7 | Gharajozd | 11.1 ± 0.95 | II | + | + | - |
| 8 | Miandoab2d | 15.2 ± 1.08 | III | + | + | T5 |
| 9 | Khankandie | 13.3 ± 1.04 | II | + | + | T4 |
| 10 | Mlalare | 12.2 ± 1.02 | II | + | + | T4 |
| 11 | Tazeghallad | 14.2 ± 0.97 | II | + | + | T4 |
| 12 | Akhtatard | 12.3 ± 1.1 | II | + | + | T4 |
| 13 | Shahrikandd | 11.5 ± 0.89 | II | + | + | T4 |
| 14 | Siminehd | 12.3 ± 1.02 | III | + | + | T4 |
| 15 | Gogjalie | 13.4 ± 1.37 | II | + | + | - |
| 16 | Gavmishgolie | 12.5 ± 0.96 | II | + | + | T4 |
| 17 | Ashigoland | 14.1 ± 1.3 | II | + | - | - |
| 18 | Gharaghshlaghe | 16.4 ± 1.76 | II | + | + | T4 |
| 19 | Hajihassane | 12.2 ± 0.88 | III | + | + | - |
| 20 | Ghanzalie | 11.7 ± 1.32 | III | + | + | T4 |
| 21 | Ghomghalad | 12.4 ± 0.97 | II | + | + | T4 |
| 22 | Vosokandd | 16.4 ± 5.02 | II,I | + | + | - |
| 23 | Ghzljad | 14.6 ± 0.99 | II | + | + | - |
| 24 | Darmand | 12.2 ± 1.2 | II | + | - | - |
| 25 | Kanikozalahe | 13.4 ± 1.03 | III | + | + | - |
| 26 | Sardarabadd | 11.6 ± 1.47 | II | + | + | T4 |
| 27 | Kanirashd | 11.1 ± 0.98 | II | + | - | - |
Morphological Observations of the Isolated Acanthamoeba spp. Amongst Positive Samples on Non-Nutrient Agar Culture Mediaa
| Source | Culture | Sample No | PCR | |
|---|---|---|---|---|
| Tap water | 0 | 16 | 0 | |
| River | Simineh Rud | 17 | 28 | 15 |
| Zarineh Rud | 10 | 16 | 9 | |
| Total (%) | 27 (45%) | 60 | 24 (40%) |
Frequency of Acanthamoeba spp. Amongst Studied Water Samples
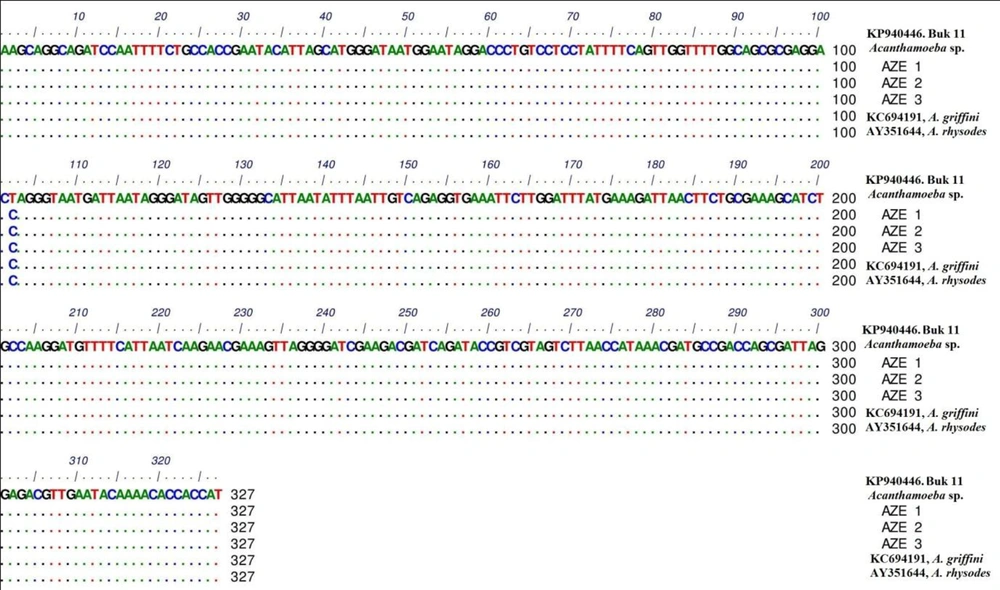
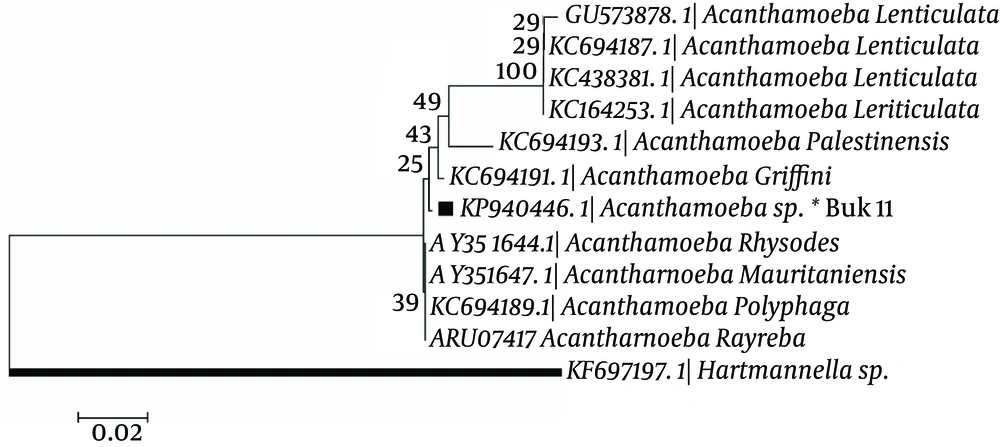
In phylogenic analysis, 14 out of 24 ampliqons were sequenced which among them, only 13 sequences (shown in Table 1) were explicitly identified as T4 genotype and one sequence was identified as T5. Twelve out of 13 sequences have common haplotypes (AZE1-AZE3: Figure 3) compared to reference isolate and only one sequence has new haplotype (Buk 11, Accession no. KP940446) which in position 101, T (Thymine) was replaced by C (Cytosine). Hartmannella spp. (KF697197) was considered as a group branch in drawing the phylogeny tree (Figure 4).
5. Discussion
This study indicated relatively high prevalence of Acanthamoeba spp. (T4) with different morphometric aspects but no more heterogeneity traits in rivers of West Azerbaijan, Northwest Iran. The high percentage of Acanthamoeba spp., in different environmental sources represents a sanitary risk for public health, particularly contact lens users and immunocompromised/suppressed patients (21).
In the present study, Acanthamoeba spp. was isolated in 45% of cultured samples, which amongst them, 88.8% were confirmed by PCR. The difference between PCR and culture method is probably due to other free living amoeba cysts except Acanthamoeba spp. Based on the morphological classification of Pussard and Pons (18), 18 cases of the positive cultures were classified in group II, with the most frequency in the world. Although, 8 positive cases were members of group III and one case from group I and group II. The results of this study showed that none of the samples from tap water contain Acanthamoeba spp., which is in contrast with some of the previous studies (4, 22). This difference, maybe, resulted from the fact that the previous studies collected their samples from a storage tank of water while our samples were obtained directly from tap water (4, 22).
Tap water is usually taken from a river or dam that it is cleaned at a purification station, such as Rand Water’s, and then supplied via pipelines to taps. Therefore, it can affect the Acanthamoeba trophozoites. Another study on the water storage tanks in UK has shown that these water supplies promote colonization of domestic water with free-living amoebae, including Acanthamoeba, and therefore increases the risk of infection with the amoebae. This accounts for the significantly greater incidence of Acanthamoeba infection in the UK and supports advice to avoid using tap water in contact lens care routines (23).
In the present study, in tap water samples the mean concentration of free residual chlorine was about 0.3 - 1 mg/L, which can be another reason that no Acanthamoeba spp. was isolated from tap water samples. It has been reported that the trophozoites of Acanthamoeba are sensitive to free residual chlorine, while the cysts are resistant to chlorine (24). On the other hand, in normal environments such as chlorine-free water, the parasite lives as trophozoite, however in frigid environments it forms cyst. In the optimum water condition, Acanthamoeba lives as a trophozoite since it is susceptible to chlorine. Therefore, it has been conceived that the growth of Acanthamoeba in water with chlorine is difficult. Given the fact that river water is chlorine-free, rivers are a significant source of the organism. In the case of the river waters, we have isolated Acanthamoeba species from 27 out of 44 (45%) river water samples by cultivation and morphologic assessment; also, 24 samples (40%) were reconfirmed by the molecular method. These results show that the contamination rate of river water in this area is very high and it maybe a result from the fact that these two rivers are exposed to air and soil, as well as other infectious sources, such as sewage, wastewater and feces of animals. These factors raise the contamination rate in the river water sources. Therefore, the residents of these areas often have an indirect contact with the Acanthamoeba infection.
In Iran, infections with Acanthamoeba spp. has increased in recent years, mainly due to poor hygiene in contact lens users (4). The ten-year study on patients with amoebic keratitis has shown that the Acanthamoeba infection is continually rising in Iran (21). Niyyati et al., from drinking and recreational water, reported Acanthamoeba in 17 (25.4%) of the 67 collected samples in East Azerbaijan, Northwest Iran. Most genotypes in this study were also reported as T4 (25).
Bagheri et al. (2010) reported that half of their investigated samples collected from cold and warm tap water sources of hospitals in 13 cities of Iran were harboring Acanthamoeba spp. (4). In another study conducted by Hooshyar et al. (2013), it is shown that 32 (80%) out of 40 water samples collected from 20 stagnant waters in squares and parks in different regions of Qazvin city, Iran, were positive for free living amoebas by the culture method. Similar to our findings, PCR reactions were positive in 14 (43.8%) out of 32 culture samples (16). Maghsood et al. (2005) isolated Acanthamoeba spp. from water samples by molecular methods in different cities of Iran (10). In the study conducted by Rezaeian et al. (2008), Acanthamoeba spp. have been recovered from 58% collected samples from a variety of ecological habitats such as tap water, soil and dust in Tehran, Iran (21).
Acanthamoeba spp. has been detected in 3% of samples taken from the James River in Virginia, USA (26). In the Pusan province, Korea, Acanthamoeba has been isolated from 5.8% of tap water samples (22). In Spain, Lorenzo-Morales et al. (2005) reported that 59.5% of tap water samples were harboring Acanthamoeba (8). The observed difference in the prevalence of Acanthamoeba in tap water in different countries might be due to the different water treatment strategies or the accuracy of the water treatment system in each country. In this study, no more new haplotypes were found in analyzed isolates, this can be associated with the structural features of 18S rRNA gene, which indicated as a conserved nucleus marker and it’s to be diploid. The findings of the present study provide the evidence for high prevalence of Acanthamoeba spp. in rivers, which reflects a risk alert for threatening human health in northwest Iran, where placed with the co-border of Iraq country. In addition, a well hygienic status of tap water considering Acanthamoeba spp. cannot be ignored in the region. This study can also serve as a platform for further explorations of water sources in Iran.