1. Background
The production of β-lactamases, including AmpC β-lactamase and extended-spectrum β-lactamase (ESBL), is an important mechanism of drug resistance in Gram-negative bacteria (1, 2). AmpC β-lactamase confers resistance to a variety of β-lactam agents, including oxyimino-cephalosporins, 7-α-methoxycephalosporins (cephamycin), monobactam, and the β-lactam/β-lactam inhibitor combination (3-6).
AmpC β-lactamase production can be chromosomal or plasmid-mediated (5, 6). Chromosomal AmpC genes are expressed constitutively at low-level mutations in the AmpC promotor/attenuator of Escherichia coli and can result in constitutive resistance. Certain other Enterobacteriaceae, such as Citrobacter spp., Serattia spp., and Enterobacter spp., carry inducible AmpC genes, which can be induced by cefoxitin and imipenem (6). However, the transfer of the AmpC gene to plasmid has resulted in dissemination among the Enterobacteriaceae, including E. coli, Klebsiella spp., Proteus mirabilis , and Salmonella spp. (7). All plasmid-mediated AmpC β-lactamase (PMABL) genes are considered clinically significant (6). Organisms such as E. coli or Klebsiella spp. that carry AmpC genes in plasmid are multidrug-resistant (5) and can cause problems for optimal clinical management and infection-control programs.
Although the exact prevalence of PMABL remains unknown in most countries, it is estimated to be lower than that of ESBL (4); although with increased frequency (8). This is due to limitations in laboratory methods for detecting PMABL. Molecular tests are required for accurate determination and confirmation (4); however, many laboratories have limited resources and prefer to achieve high pretest probability before performing polymerase chain reaction (PCR) analyses (9). Reduced susceptibility to cefoxitin has been an indicator for probable AmpC production (3, 4, 7, 9, 10) but it is non-specific and can be mediated by alterations in outer membrane permeability (7). Some authors also suggest that isolates with non-susceptibility to ESBL cephalosporins should be suspected to carry AmpC genes (9).
While plasmid-mediated AmpC causes clinical concern for the improvement of the clinical management of infection and infection-control measures (7), the available phenotypic tests are not convenient and lack sufficient sensitivity and specificity (1). Presently, there is no standard phenotypic test to identify these organisms. The current Clinical and Laboratory Standard Institute (CLSI) guidelines do not describe any method for the detection of AmpC β-lactamase (8).
2. Objectives
The present study was designed to determine the prevalence of plasmid-mediated AmpC-producing isolates of E. coli, Klebsiella spp. and P. mirabilis with reduced susceptibility to cefoxitin or extended-spectrum cephalosporins, using the multiplex PCR method.
3. Methods
A total of 310 consecutive non-duplicate isolates of E. coli, Klebsiella spp. and P. mirabilis obtained from various clinical specimens, including blood, wounds (pus), urine, and respiratory tract excretions, were received in the microbiology laboratory of Shariati Hospital, affiliated with Tehran University of Medical Sciences, from January 1, 2015, to April 1, 2015. The isolates were identified by conventional microbiological procedures. Table 1 summarizes the number and percentage of isolates from the different specimen types. Antibiotic susceptibility was determined using the Kirby-Bauer disk diffusion method according to CLSI guidelines (11).
| Isolate | Specimen Type | ||||||
|---|---|---|---|---|---|---|---|
| Urine | Blood | Body Fluid | Respiratory Excretion | Wound and Pus | Catheter | Total | |
| E. coli | 163/206 (79.13%) | 8/14 (57.14%) | 18/19 (94.74%) | 11/21 (52.38%) | 14/38 (36.84%) | 7/12 (58.33%) | 221/310 (71.29%) |
| Klebsiella spp. | 33/206 (16.02%) | 4/14 (28.57%) | 1/19 (5.26%) | 10/21 (47.62%) | 24/38 (63.16%) | 5/12 (41.67%) | 77/310 (24.84%) |
| P. vulgaris | 10/206 (4.85%) | 2/4 (14.29%) | 0/19 (0.00%) | 0/21 (0.00%) | 0/38 (0.00%) | 0/12 (0.00%) | 12/310 (3.87%) |
Frequency of Isolates with Regard to Specimen Type
Screening for AmpC β-lactamase production was done by placing a cefoxitin disk (30 µg; Rosco, Denmark) on Mueller-Hinton agar (2, 3, 8, 12). Isolates showing an inhibition zone diameter of < 18 mm were considered positive on the screening test and were subjected to further molecular evaluation.
Screening and confirmation of ESBL production was also performed by the disk diffusion method as per the CLSI, using ceftazidime 30 µg, cefotaxime 30 µg, and ceftriaxone 30 µg disks for screening, followed by combined disks of ceftazidime (30 µg)/ceftazidime-clavulanate (30 µg/10 µg) and cefotaxime (30 µg)/cefotaxime-clavulanate (30 µg/20 µg) for confirmation (Rosco, Denmark) (11). Isolates with positive screen tests that failed to be confirmed as ESBL by the combined disk method were also considered for further molecular testing.
The genomic DNA of the isolates with positive screening tests for AmpC was extracted using extraction kits (Bioneer, Seoul, Korea) according to the manufacturer’s instructions, followed by multiplex PCR assays to detect four family-specific AmpC genes carried on plasmid, including FOX, MOX, DHA, and CIT. PCR was carried out with final volume of 25 µL (Eppendorf thermocycler; Germany). The primers (13) used for PCR assay are listed in Table 2.
| Target Gene | Primer | Sequence (5’ to 3’, as Synthesized) | Amplicon Size (bp) |
|---|---|---|---|
| MOX-1, MOX-2, CMY-1, CMY-8 to CMY-11 | MOXMF | GCT GCT CAA GGA GCA CAG GAT | 520 |
| MOXMR | CAC ATT GAC ATA GGT GTG GTG C | ||
| LAT-1 to LAT-4, CMY-2 to CMY-7, BIL-1 | CITMF | TGG CCA GAA CTG ACA GGC AAA | 462 |
| CITMR | TTT CTC CTG AAC GTG GCT GGC | ||
| DHA-1, DHA-2 | DHAMF | AAC TTT CAC AGG TGT GCT GGG T | 405 |
| DHAMR | CCG TAC GCA TAC TGG CTT TGC | ||
| FOX-1 to FOX-5b | FOXMF | AAC ATG GGG TAT CAG GGA GAT G | 190 |
| FOXMR | CAA AGC GCG TAA CCG GAT TGG |
Primers Used for Characterization of AmpC β-lactamase
The cycling conditions were as follows: initial DNA release and denaturation at 94°C for 3 min followed by 35 cycles at 94°C for 1 minute, 53°C for 1 minute, and 72°C for one minute, with a final extraction at 72°C for 5 minutes. The amplicons were analyzed by gel electrophoresis (agarose gel), stained with ethidium bromide, and visualized by UV transillumination. Standard E. coli (ATCC25922) and Klebsiella pneumoniae (ATCC 700603), or previously known positive and negative isolates of E. coli, were included in each run.
4. Results
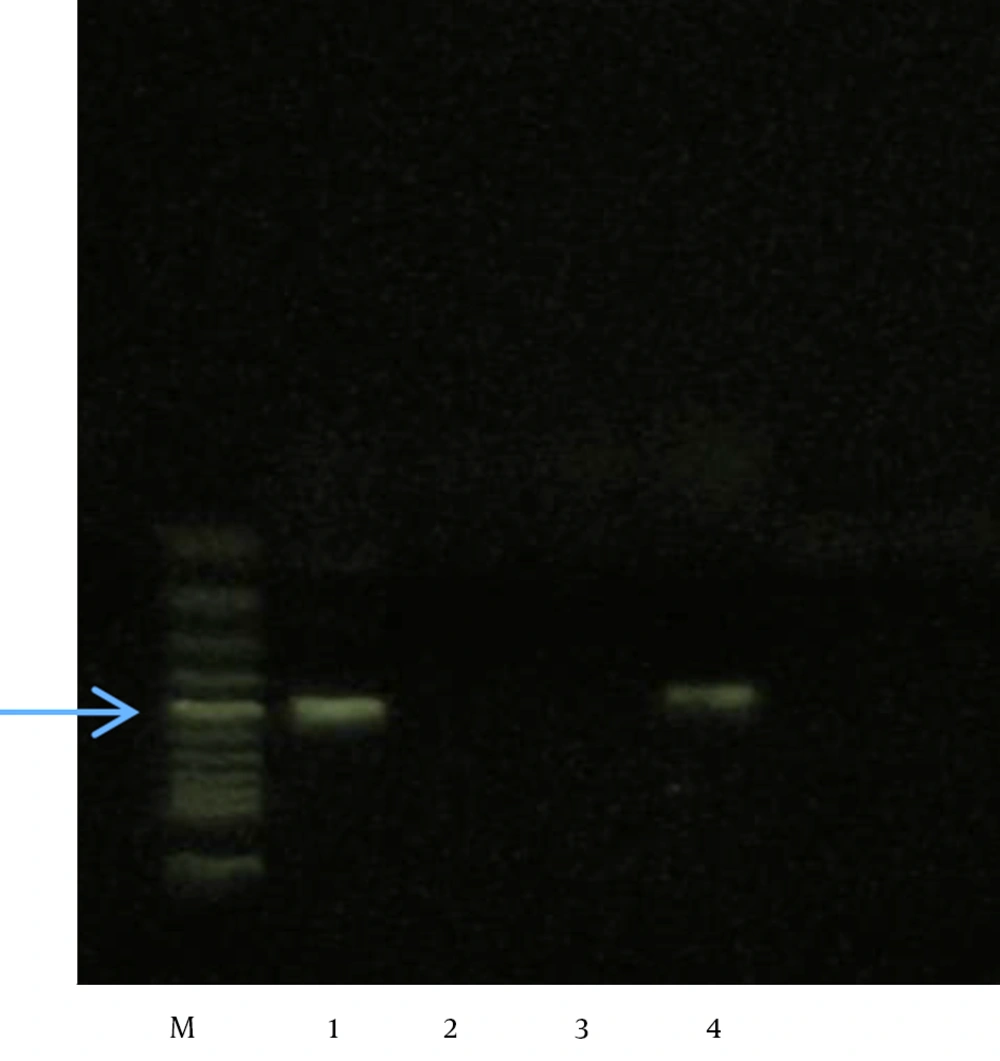
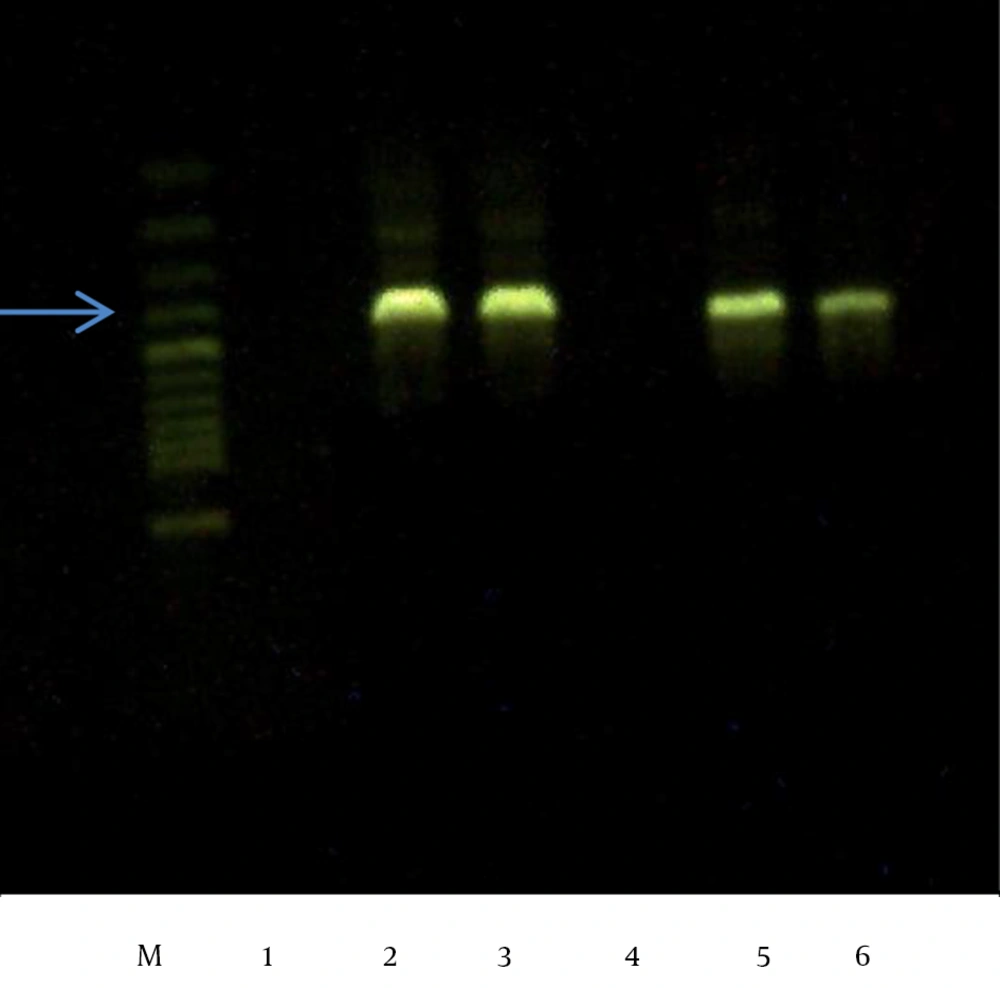
A total of 310 isolates were identified as E. coli (n = 221), Klebsiella spp., (n = 77), or P. mirabilis (n = 12). Among these, 50 isolates that showed inhibition zones of < 18 mm on the cefoxitin disk and/or had positive ESBL screening tests but failed to be confirmed were all subjected to PCR analysis. Of these 50 isolates, 47 showed a positive reaction (94%), including 16 (34.04%) for CIT only, 7 (14.89%) for DHA only, and 24 (51.06%) for both DHA and CIT. No isolate was positive for FOX or MOX (Figures 1 and 2)
The frequency distribution of plasmid-mediated AmpC and ESBL-producing isolates with regard to different medical wards and clinical specimens is summarized in Tables 3 and 4, respectively. Medical wards were categorized as emergency medicine (general, oncology, obstetrics), internal medicine (nephrology, respiratory tract, general, hematology/oncology, bone marrow transplant, renal transplant, endocrinology, gastrointestinal tract, heart and coronary care unit, surgical (general, orthopedics, urology, neurosurgery, gynecology, head and neck), and intensive care (general, neonatal, neurosurgical).
| Isolate | Hospital Ward | ||||
|---|---|---|---|---|---|
| Emergency Department | Internal Medicine Wards | Surgery Wards | Intensive Care Units | Total | |
| Plasmid-mediated AmpC producer | 21/143 (14.68%) | 12/79 (15.18%) | 4/36 (11.11%) | 10/52 (19.23%) | 47/310 |
| ESBL-producing organisms | 68/143 (47.55%) | 44/79 (55.69%) | 16/36 (44.44%) | 24/52 (46.15%) | 152/310 |
Frequency of AmpC- and ESBL-producing isolates with regard to different wards
| Isolate | Specimen Type | ||||||
|---|---|---|---|---|---|---|---|
| Urine | Blood | Body Fluid | Respiratory Excretion | Wound and Pus | Catheter | Total | |
| Plasmid-mediated AmpC producer | 29/206 (1.15%) | 2/14 (14.28%) | 3/19 (15.78) | 4/21 (19.04%) | 8/38 (21.05%) | 1/12 (8.33%) | 47/310 |
| ESBL-producing organisms | 116/206 (56.31%) | 8/14 (57.14%) | 12/19 (63.15%) | 6/21 (28.57%) | 10/38 (26.31%) | 0/12 (0.00%) | 152/310 |
Frequency of AmpC- and ESBL-producing isolates with regard to different sample types
The greatest number of AmpC-producing organisms were recovered from emergency departments (21/47); however, considering the number of submitted specimens, the intensive care units showed the highest prevalence of AmpC-producing organisms (19.23%), while surgical wards revealed the lowest frequency (11.11%). The specimen type most often positive for AmpC-producing organisms was urine (29/47). However, again considering the number of samples, the highest percentage came from wound cultures (8/38, 21.05%).
Positive PCR reactions were identified in 35/221 (15.83%) and 12/77 (15.58%) of the E. coli and Klebsiella spp. isolates, respectively. None of the Proteus organisms were positive for the AmpC gene. The frequencies of different genes are summarized in Table 5. Of the total of 310 organisms, 86/221, 64/77, and 2/12 isolates of E. coli, Klebsiella spp., and P. vulgaris, respectively, were positive for ESBL production. Twenty-nine and 7 isolates of E. coli and Klebsiella spp., respectively, were positive for both ESBL and AmpC production. All of the isolates were susceptible to imipenem.
| Isolate | Gene | |||||
|---|---|---|---|---|---|---|
| CIT | DHA | CIT-DHA | FOX | MOX | Total | |
| Klebsiella spp. | 3 | 4 | 5 | 0 | 0 | 12 |
| E. coli | 13 | 3 | 19 | 0 | 0 | 35 |
Frequency of AmpC genotypes in clinical isolates
5. Discussion
Plasmid-mediated AmpC β-lactamases are becoming more important clinically (9), and their recognition will be beneficial for both surveillance and for epidemiological measures and infection control (2), in order to avoid nosocomial outbreaks and treatment failures (6). Enzyme-extraction methods have been introduced for the phenotypic detection of AmpC activity (7); however, these are not suitable for routine clinical use (7). Various inhibitory-based methods, such as boronic acid compounds, cloxacillin, M3D (5, 7, 8), a modified double-disk test (5, 8, 14), and an AmpC disk test (5), have also been employed. However, as mentioned above, these methods have limited sensitivity and specificity (1). Moreover, they are unable to differentiate between chromosomal and plasmid-mediated AmpC producers (7). Therefore, molecular tests are still required for reliable identification of organisms carrying AmpC genes on plasmid (4).
Using multiplex PCR as a confirmatory test in the present study, the prevalence of plasmid-mediated AmpC genes in isolates of E. coli, Klebsiella spp., and P. mirabilis was 15.16%. A 2004 report from the United States documented that 7% - 8.5% of Klebsiella spp. and 4% of E. coli isolates contained plasmid-mediated AmpC genes (4, 15). In Asia, using different screening and confirmatory methods, the prevalence of plasmid-mediated AmpC is variable. A study conducted in northern Iran reported a rate of 7.7% based on phenotypic methods (16). In another study in Tehran, 10.2% of E. coli isolates were AmpC-positive according to the PCR method (17). The lower prevalence compared to our results could be due to differences in PCR assay design, as the abovementioned studies did not apply primers for detecting DHA and MOX genes in the assay protocol (17). Peter-Getzlaff et al. (6) reported that 21/51 (41%) of E. coli isolates in their study were AmpC producers, among which 9 (19.6%) carried plasmid-mediated genes (6). In another study from India, 312/909 isolates were positive on the cefoxitin screening test, of which 114 (36.5%) were confirmed on PCR (4); however, the prevalence of plasmid-mediated AmpC was approximately 12.5%, which is rather similar to our findings.
In the present study, the prevalence rates of PMABL in E. coli and Klebsiella spp. isolates were 15.83% and 15.58% respectively, which are quite similar. In a study by Shafiq et al. (2) in Pakistan, of 55/103 positive-screen AmpC-producing isolates, 7.9% of E. coli and 12.37% of Klebsiella isolates carried AmpC genes. In another study in Pakistan (2, 18), the rates were 18% and 14% in isolates of E. coli and Klebsiella, respectively, which were closer to our findings. However, Fam et al. (10) reported that the prevalence of plasmid-mediated AmpC genes were significantly higher in Klebsiella isolates compared to E. coli (43.5% versus 17.7%) (10). It has been suggested that some PMABLs may be clinically more important (1). In a study by Black et al. (1), the inducible DHA-1 enzyme and the constitutively produced XMY-1-like enzymes were associated with 46% versus 14.3% mortality rates, respectively. However, the authors mentioned that due to the small size of the specimens, the significance of their findings should be further investigated (1).
In our study, genes belonging to the CIT family were more common in E. coli (n = 13) than in Klebsiella spp. (n = 3), while for DHA, the number was rather equal (n = 3 and n = 4, respectively). However, the most prevalent genotype in both types of isolates was CIT-DHA. In a study by Mohamudha et al. (5), DHA was more common in both Klebsiella spp. and E. coli isolates (46.7% and 38%, respectively). In another report, all E. coli isolates were positive for the CIT family (17) and no FOX was detected. Fam et al. (10) and Manoharan et al. (4) found that the CIT and CIT-FOX genes were more frequent. In other studies as well, no AmpC producers carried FOX or MOX (7, 10). The ACC gene also appears to be uncommon according to different studies (4, 5, 7, 10). The latter is important because using cefoxitin disks as a screening method is unable to detect isolates producing plasmid-encoded AmpC β-lactamase of the ACC family (6). Our study was limited due to a lack of primers able to amplify genes belonging to the ACC family, preventing us from evaluating the ACC genotype status in our isolates.
Ten out of 52 (19.23%) of the isolates from ICUs carried plasmid-mediated AmpC genes. This is unsurprising because patients can be compromised or exposed to previous cephalosporin therapy. However, the most specimens were isolated from the emergency departments (21/47, 44.6%), mainly due to referral cases admitted in our hospital. This needs special attention because it may indicate the spread of PMABL-producing strains in the community, as some other studies have suggested (2, 8). The prevalence of ESBL-producing isolates was 49% in our study, which is a little lower than the rates of 57% and 52% previously reported in Iran (16, 17).
Plasmid-mediated AmpC genes have the capacity to transfer and spread to other organisms within hospital settings, leading to nosocomial infections and treatment failures. To date, phenotypical tests are not able to accurately and reliably recognize PMABL organisms. Although not feasible for routine testing, clinical laboratories, especially in referral centers, should employ molecular testing for surveillance studies.