1. Background
Staphylococcus aureus is one of the most common and important pathogens, accounting for diverse nosocomial and community acquired infections. The bacterium has potentiality to cause three types of disorders in human: superficial, toxin mediated, and lethal-systemic infections (1). The serious concern about this bacterium is development of antibiotic resistance, especially against methicillin, the so-called methicillin resistant S. aureus (MRSA) (2, 3). The emergence of MRSA has left with very few antibiotic alternatives to treat S. aureus-related infections, which has caused renewed interest in the usage of macrolide, lincosamide, and streptogramin B (MLSB) antibiotics against S. aureus infections (2, 4). Though these antibiotics differ structurally, they act in a similar way. They inhibit protein synthesis via binding to large subunit of ribosome (4, 5). Among these antibiotics, clindamycin is preferred because of its excellent pharmacokinetic properties (6, 7). Unfortunately, mistreatments with MLSB antibiotics have caused an unusual increase in the rate of resistance to these antibiotics leading to clinical treatment failure (8).
Three main mechanisms have been elucidated for MLSB antibiotics resistance in staphylococci: target site modification by a methylase enzyme encoded by erm genes, including ermA, ermB, ermC, or ermF (ermA and ermC are more common); Macrolide efflux pump encoded by msrA or msrB genes; and inactivation of lincosamides due to lincosamide nucleotidyl transferase enzyme which is encoded by inuA gene (9, 10).
In vitro, S. aureus isolates with inducible resistance are resistant to erythromycin but appear susceptible to clindamycin. In this situation, therapy with clindamycin may be selective for constitutive erm mutants, which can lead to clinical treatment failure (6, 8); but, erythromycin-resistant staphylococci should not be assumed as clindamycin-resistant (11). Since isolates with msrA-mediated efflux also appear erythromycin resistant and clindamycin susceptible in in-vitro tests, such isolates do not typically become clindamycin resistant during therapy (11, 12).
Unlike constitutive (c) MLSB (cMLSB) resistance, inducible (i) MLSB (iMLSB) resistance is not recognized in conventional laboratory tests (8, 9). Inaccurate identification of iMLSB resistance may lead to clinical failure of clindamycin therapy; conversely, labeling all erythromycin-resistant staphylococci as clindamycin resistant prevents the use of clindamycin in infections caused by truly clindamycin susceptible staphylococcal isolates (6, 8, 11).
The incidence of MLSB resistance varies significantly according to geographical region, from hospital to hospital, and age group (6, 12). On the other hand, the frequency of iMLSB in the north west of Iran has not been studied thoroughly. By using a simple disk approximation test, namely D-test, microbiology laboratories can differentiate isolates which are iMLSB resistant and harbor erm genes from those that are truly sensitive to clindamycin and show efflux pump mediated resistance as macrolide and streptogramin B (MSB) or negative phenotype (due to msrA gene) (6, 8, 9, 11, 12).
2. Objectives
The present study investigated the frequency of MLSB resistance among clinical isolates of S. aureus isolated from various teaching hospitals in the north west of Iran, including MRSA and methicillin susceptible S. aureus (MSSA) by phenotypic and genotypic methods and assessed resistance to therapeutic agents which are recommended for these isolates.
3. Methods
3.1. Isolation and Identification of Staphylococcus aureus
During a period of one year from February 2014 to March 2015, all clinical specimens submitted to microbiology laboratories of educational health care centers of Tabriz University of Medical Sciences in the Northwest of Iran were screened for presence of S. aureus isolates. Repetitive isolates from the same patient were not examined in this study. The isolates were obtained from different specimens of inpatients and outpatients such as: wound, blood, urine, abscess, fistula and catheter aspirates, sputum and other body fluids and were confirmed by standard microbiology tests (13). They were further confirmed by PCR assay for the presence of nuc gene in all the isolates (14). Two hundred and fifteen isolates were collected and stored in TSB broth supplemented by 30% glycerol at -70°C.
3.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility test was performed as per the clinical and laboratory standards institute (CLSI-2014) guidelines (15), with a panel of following antibiotics (MAST, Group Ltd, Merseyside, UK): oxacillin (1 µg), cefoxitin (30 μg), penicillin (10 U), gentamicin (10 µg), erythromycin (15 µg), clindamycin (2 µg), rifampin (30 µg), ciprofloxacin (5 µg), trimetoprim-sulfametoxazol (1.25/23.75 μg), and linezolid (30 µg). The minimal inhibitory concentrations (MICs) were determined by E-test on Mueller-Hinton’s agar plates (MHA, Merck, Germany) for vancomycin according to the manufacturer’s recommendations (Liofilchem, Italy) and the breakpoints for resistance were those as defined by the CLSI-2014. Staphylococcus aureus ATCC 25923, S. aureus ATCC 29213, S. aureus ATCC 33591, Enterococcus faecalis ATCC 29212, and E. faecalis ATCC 51299 were used as the control strains.
3.3. Disk Approximation Test (D-Test)
D-zone test was performed as per the CLSI 2014 guidelines (15). Briefly, a suspension of overnight growth of isolates equivalent to 0.5 McFarland turbidity was inoculated on MHA plates. For detecting inducible clindamycin resistance, 15 μg erythromycin disks and 2 μg clindamycin disks (MAST, Group Ltd, Merseyside, UK) were placed on MHA plates (15 mm edge to edge) as part of a standard disk diffusion test. Following overnight incubation at 35°C, inducible clindamycin resistance was observed as flattening of zone towards clindamycin (D-shape), which indicates the isolate has inducible clindamycin resistance (because of erm gene) (9). No flattening of zone towards clindamycin indicates the isolate is erythromycin resistant only (due to msrA gene). As an isolate demonstrated inducible resistance, clindamycin was reported as resistant (6, 9). Based on D-test results, S. aureus isolates were categorized into four non-inducible and two inducible resistance phenotypes according to a previous study (9).
3.4. DNA Extraction
Briefly, DNA was extracted using dodecyl sulphate sodium salt (SDS, Merck, Germany) - proteinase K (CinnaGene, Tehran, Iran) method modified with N-cetyl-N, N, N-trimethyl ammonium bromide (CTAB, Merck, Germany) (3). The concentration of extracted DNA was confirmed by Nano drop 1000 (NanoDrop, Wilmington, USA).
3.5. Identification of nuc, mecA, erm and msrA Genes by PCR
Confirmation of all S. aureus isolates was carried out by PCR for amplification of nuc gene (279 bp) using primer pair (CinnaGene, Tehran, Iran, nuc/F: 5’-GCGATTGATGGTGATACGGTT-3’ and nuc/R: 5’-AGCCAAGCCTTGACGAACTAAAGC-3’) (14). PCRs were performed in a final volume of 25 μL with an automated thermal cycler (Eppendorf mastercycler gradient, Germany) with the PCR cycling conditions which were as follows: initial cycle at 94°C for 5 minute, followed by 37 cycles at 94°C for 1 minute, 55°C for 30 seconds, 72°C for 1 minute and 30 seconds, and final extension cycle at 72°C for 3 minutes and 30 seconds. PCR products were analyzed on 1.5% agarose gel, which was visualized under ultraviolet illumination (Gel documentation, UVP, UK). Negative controls for each used primer contained all the components except template DNA. Staphylococcus aureus ATCC 25923 and S. epidermidis ATCC 12228 were used as positive and negative control strains, respectively.
Staphylococcus aureus isolates were confirmed as MRSA based on the presence of mecA gene (310 bp) among our isolates of S. aureus using primer pair (CinnaGene, Tehran, Iran, mecA/F: 5’- GTAGAAATGACTGAACGTCCGATAA-3’ and mecA/R: 5’-CCAATTCCACATTGTTTCGGTCTAA-3’) (3). PCR cycling conditions were as follows: initial cycle at 94°C for 4 min, followed by 30 cycles at 94°C for 45 seconds, 56°C for 45 seconds, 72°C for 1 minute, and final extension cycle at 72°C for 7 minutes. Staphylococcus aureus ATCC 33591 and S. aureus ATCC 25923 were used as positive and negative control strains, respectively.
Amplification of ermA (139 bp), ermB (142 bp), ermC (190 bp), and msrA (163 bp) genes was carried out according to previous studies (16, 17). Oligonucleotide primers used for PCR were as follows: ermA/F: 5’-TATCTTATCGTTGAGAAGGGATT-3’, ermA/R: 5’-CTACACTTGGCTTAGGATGAAA-3’, ermB/F: 5’-CTATCTGATTGTTGAAGAAGGATT-3’, ermB/R: 5’-TTTACTCTTGGTTTAGGATGAAA–3’, ermC/F: 5’-CTTGTTGATCACGATAATTTCC-3’, ermC/R: 5’-ATCTTTTAGCAAACCCGTATTC-3’, msrA/F: 5’-TCCAATCATTGCACAAAATC-3’, msrA/R: 5’-AATTCCCTCTATTTGGTGGT-3’. PCR conditions were as follows: Initial denaturation at 95°C for 3 minutes; followed by 35 cycles at 95°C for 30 seconds, various annealing temperatures (62.8°C for ermA, 59°C for ermB, 58°C for ermC, and 55°C for msrA) for 30 seconds, followed by extension at 72°C for 45 seconds and final extension at 72°C for 7 minutes. Staphylococcus aureus isolates with ermA, ermC, ermB, and msrA genes and S. aureus strains (ATCC 25923 and ATCC 29213) were used as positive and negative control strains, respectively.
3.6. Statistical Analysis
Data were analyzed by Chi-square test using SPSS 22.0 statistical software (SPSS Inc., Chicago, IL). Astatistically significant difference was considered as P value < 0.05.
3.7. Ethical Approval
This work was approved by the ethics committee of Tabriz University of Medical Sciences (reference No. 5/4/3978).
4. Results
A total of 215 isolates of S. aureus were collected from various clinical specimens and identified by standard tests.
4.1. Molecular Tests
The nuc gene (279 bp) was amplified by all of our isolates. PCR for mecA gene was performed on S. aureus isolates that showed 87 (40.5%) as MRSA and 128 (59.5%) as MSSA. The distribution of MRSA isolates was as follows: 63 (72.41%) from in-patients and 24 (27.59%) from out-patients (P = 0.649), while for MSSA isolates it was 89 (69.53%) from in-patients and 39 (30.47%) from out-patients (P = 0.649).
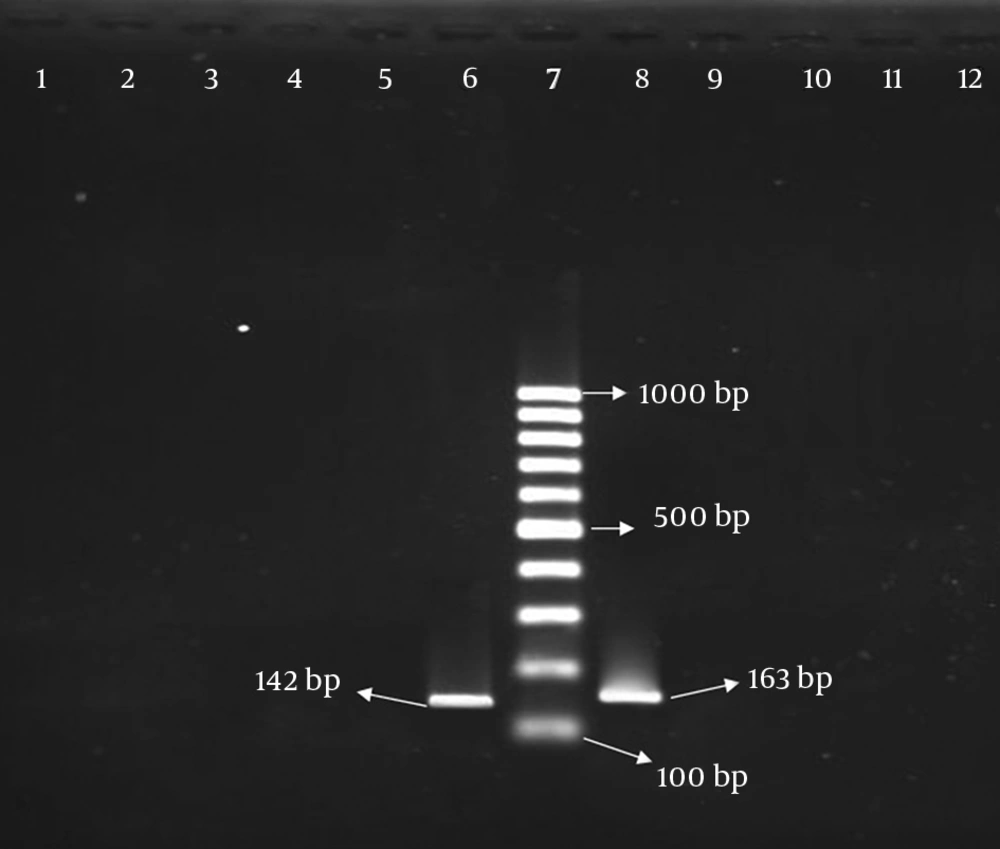
According to PCR results, 46 (21.40%) and 84 (39.06%) S. aureus isolates were positive for presence of ermA and ermC genes, respectively. Out of 215 isolates, thirty nine (18.14%) showed both ermA and ermC genes. Although 6 (2.79%) isolates were positive on D test, they were negative for the presence of erm genes. All isolates were negative for the presence of ermB and msrA genes (Figure 1). The distribution of above-mentioned genes among MRSA and MSSA isolates is shown in Table 1.
Lanes 1 to 3, isolates without ermB gene; lane 4, reagent control; lane 5, S. aureus ATCC 25923 as negative control, lane 6, control isolate with ermB gene; lane 7, Size marker 1000 bp DNA Ladder; lanes 8, control isolate with msrA gene; lane 9, S. aureus ATCC 29213 as negative control; lanes 10 to 12, isolates without msrA gene.
| Genes | MRSA (n = 87) | MSSA (n = 128) | Total isolates (n = 215) |
|---|---|---|---|
| ermA alone | 44 (50.57) | 2 (1.56) | 46 (21.40) |
| ermC alone | 67 (77.01) | 17 (13.28) | 84 (39.06) |
| ermA + ermC | 38 (43.67) | 1 (0.78) | 39 (18.14) |
| Without ermA and ermC | 4 (4.60) | 2 (1.56) | 6 (2.79) |
aValues are expressed as No. (%).
bNo ermB and msrA genes were detected in the present study.
4.2. Phenotypic Tests
The results of antimicrobial sensitivity test showed that all isolates (100%) were susceptible to linezolid and the majority of them (96.3%) were resistant to penicillin. The results of antibiotic susceptibility testing for other antibiotics are shown in Table 2. The rates of resistance among MRSA and MSSA isolates against studied antibiotics were as follows: penicillin, 100% and 93.75% (P = 0.017); oxacillin, 89.7% and 0% (P = 0.000); cefoxitin, 96.6% and 0% (P = 0.000); clindamycin, 88.5% and 15.62% (P = 0.000); erythromycin, 86.2% and 15.62% (P = 0.000); ciprofloxacin, 83.9% and 3.12% (P = 0.000); gentamicin, 83.9% and 0.78% (P = 0.000); trimethoprim-sulfamethoxazole, 55.17% and 1.56% (P = 0.000); rifampin, 39.08% and 0% (P = 0.000), respectively. Antibiotic resistance patterns of 87 MRSA isolates are shown in Table 3. The isolates fell into 10 distinctive antibiotic resistance profiles.
| Antibiotics | S | I | R |
|---|---|---|---|
| Penicillin | 8 (3.7) | - | 207 (96.3) |
| Oxacillin | 137 (63.7) | - | 78 (36.3) |
| Cefoxitin | 131 (60.93) | - | 84 (39.07) |
| Gentamicin | 141 (65.6) | - | 74 (34.4) |
| Erythromycin | 120 (56) | - | 95 (44) |
| Clindamycin | 118 (55) | - | 97 (45) |
| Trimethoprim-sulfamethoxazol | 165 (76.7) | - | 50 (23.3) |
| Ciprofloxacin | 138 (64.2) | - | 77 (35.8) |
| Rifampin | 181 (84.2) | - | 34 (15.8) |
| Linezolid | 215 (100) | - | 0 (0) |
Abbreviations: I, intermediate; R, resistant; S, sensitive.
| Resistance Patterns | No. (Identity of Isolates) | Antibiotic Resistance Patterns | MDRa or Non-MDR |
|---|---|---|---|
| R1 | 4 (14, 75, 111, 119) | P, OX, FOX | Non-MDR |
| R2 | 2 (132, 135) | P, OX, FOX, GM, CC, SXT | MDR |
| R3 | 39 (2 - 5, 8, 15, 25, 37, 41, 42, 44, 47, 61-63, 91, 98, 107, 114, 117, 118, 123, 124, 136, 140, 151, 152, 154, 156-158, 160, 162, 173, 175, 176-178, 209) | P, OX, FOX, GM, CC, E, CP, SXT | |
| R4 | 26 (10, 11, 13, 16, 19, 22, 28, 40, 48, 57, 76, 83, 96, 97, 105, 153, 155, 159, 161, 174, 181, 182, 201-203, 206) | P, OX, FOX, GM, CC, E, CP, R | MDR |
| R5 | 6 (60, 68, 74, 78, 125, 179) | P, OX, FOX, GM, CC, E, CP, SXT, R | MDR |
| R6 | 1 (192) | P, OX, FOX, CC, E, CP, R | MDR |
| R7 | 1 (7) | P, FOX, CC, E, CP, SXT | MDR |
| R8 | 2 (20, 147) | P, FOX, CC, E | MDR |
| R9 | 3 (109, 115, 150) | P, FOX | Non-MDR |
| R10 | 3 (39, 50, 51) | P | Non-MDR |
Abbreviations: CC, clindamycin; CP, ciprofloxacin; E, erythromycin; FOX, cefoxitin; GM, gentamicin; OX, oxacillin; P, penicillin; R, rifampin; SXT, trimethoprimsulfamethoxazole.
aMultidrug-resistant (isolates that are resistant to more than 3 antimicrobial agents, 88.50% of the MRSA isolates are MDR).
Seventy seven out of 87 MRSA isolates (88.50%) were resistant to more than 3 antimicrobial agents, and thus they were recorded as multidrug-resistant (MDR) (Table 3). The MIC of isolates against vancomycin was in the range of 0.25 μg/mL to 6 μg/mL and the MIC50 and MIC90 for the isolates were 0.5 and 1 μg/mL, respectively. Only 3 MRSA isolates, which showed MICs equal to 6 μg/mL, were recorded as vancomycin intermediate-resistant S. aureus (VISA). MRSA isolates were resistant to antibiotics significantly more than MSSA (P = 0.000) except for penicillin, linzeolide, and vancomycin. All MRSA and MSSA isolates were susceptible to linzeolide (P = 0.409) and vancomycin (P = 0.409, except 3 MRSA isolates).
Based on D test results, various phenotypes detected among 215 isolates of S. aureus are shown in Table 4. Table 5 depicts the rate of inducible clindamycin resistance (ICR) in MSSA and MRSA isolates. Though this rate was higher in MSSA (11.71%) than MRSA (9.19%) isolates, this difference was not meaningful (P = 0.557). In contrast, the rate of constitutive MLSB resistance was significantly higher in MRSA (79.31%) than MSSA (3.90%) isolates (P = 0.000). MSB phenotype was not detected in this study.
| D Test Phenotypes | Resistance Phenotypes | CC Result | E Result | S. aureus (215 Isolates) | D Test Description |
|---|---|---|---|---|---|
| D | Inducible MLSB | S | R | 13 (6.04) | D shaped clear zone around CC disc proximal to E disc |
| D+ | Inducible MLSB | S | R | 10 (4.65) | |
| D- | MS or Neg. | S | R | 0 | Clear zone only around CC disc |
| R | Constitutive MLSB | R | R | 71 (33.02) | Growth upto CC and E discs |
| HD | Constitutive MLSB | R | R | 3 (1.39) | Two zones of growth around CC disc |
| S | No resistance | S | S | 118 (54.88) | Clear zone around both discs |
Abbreviations: CC, clindamycin; D, D zone test, E, erythromycin; HD, hazy D zone; Neg, Negative; R, resistant; S, sensitive.
aValues are expressed as No. (%).
| Isolate Name | Total Isolates | Constitutive MLSB | Inducible MLSB | S Phenotype |
|---|---|---|---|---|
| S. aureus | 215 | 74 (34.42) | 23 (10.69) | 118 (54.88) |
| MSSA | 128 (59.5) | 5 (3.90) | 15 (11.71) | 108 (84.37) |
| MRSA | 87 (40.5) | 69 (79.31) | 8 (9.19) | 10 (11.49) |
aValues are expressed as No. (%).
Our findings revealed that 75 (86.20%) of MRSA isolates were resistant to erythromycin among whom 66 (88%) and 9 (12%) isolates were cMLSB and iMLSB, respectively. On the other hand, 20 (15.62%) of MSSA isolates were resistant to erythromycin, and cMLSB and iMLSB phenotypes were seen in 5 (25%) and 15 (75%) isolates, respectively. Among 152 in-patients and 63 out-patients, ICR phenotype was observed in 14 (9.21%) and 9 (14.28%) isolates, respectively (P = 0.273).
5. Discussion
The emergence of VISA is a great concern and also an alarm for clinicians to give a second thought to the usage of this antibiotic (vancomycin) or finding an alternative treatment such as MLSB antibiotics (2, 4-6, 8, 9, 17, 18). Unfortunately, misuses of MLSB antibiotics have led to an unusual increase in the rate of resistance to these antibiotics especially to clindamycin (8, 9). Therefore, it is important for microbiology laboratories to correctly recognize and report whether an S. aureus isolate is truly clindamycin susceptible or not. This true result can be obtained by using a simple disk agar diffusion test, described as D-zone test, because this test can exclude inducible clindamycin resistance (8, 9, 11, 12, 17).
In the present study, the prevalence of iMLSB, cMLSB, MSB, and S phenotypes among all the S. aureus isolates was 10.69%, 34.42%, 0%, and 54.88%, respectively. The frequency of ICR was in agreement with previous findings from Iran and India (19-22). However, lower rates of ICR (5.2%, 5.3%, and 8.64%) were reported by other researchers (23-25). In contrast to our finding, the higher rates of ICR have been reported (20.3%, 20.7%, 32.3%, and 33.3%) by other investigators (17, 26-28). Such differences in the ICR pattern could be due to differences in prescriptions of MLSB drug groups.
In contrast to many studies, our finding in this study showed that the frequency of inducible resistance phenotype was higher in MSSA (11.71%) than MRSA (9.19%) isolates (P = 0.557) (10, 17, 23-26). Similar to our study, in a study from southeastern of Turkey it was shown that inducible clindamycin resistant strains were more prevalent in MSSA (10%) than MRSA (6.9%), nevertheless, this difference was also not significant (P = 0.434) (29).
In the present study, constitutive clindamycin resistance was seen in 74 (34.42%) S. aureus isolates that was comparable with two studies from Iran and India (25, 30). Other researchers have reported either much lower prevalence (12.9%, 16.6%, and 23.3%) or higher rates (36%, 37.5% and 40%) (17, 19, 23, 27, 31, 32). Our finding shows the prevalence of constitutive clindamycin resistance phenotype was 79.31% in MRSA and 3.90% in MSSA isolates (P = 0.000). This predominance has also been reported by most studies (20, 29). The reasons of above undulations among various reports are the MLSB resistance pattern which varies widely among geographical region, age, source and type of strains, susceptibility to methicillin, and even among medical centers, and as previously mentioned, such differences could be due to differences in the form of drug usage (6, 9, 11, 12).
Hazy D (HD) phenotype was detected in 3 (3.45%) MRSA isolates. This type of resistance must be considered as R phenotype and its rate has been reported rarely in different countries (10). MSB or negative phenotype was not found in our study and all the erythromycin resistant and clindamycin susceptible isolates showed inducible resistance phenotype. The rates of MSB phenotype among S. aureus isolates have been reported to vary from 5.7% to 44.8% in other countries (19-21, 25-28, 31). These differences in the rates of MSB phenotype which is related to msrA genes emphasize the importance of performing D-test for differentiation of truly clindamycin susceptibility from iMLSB phenotype and selecting proper therapeutic agent.
In this present study, ermA and ermC genes were observed in 46 (21.40%) and 84 (39.06%) isolates, respectively, while much higher frequencies have been reported in other Iranian studies (60.3% - 54.8% and 41.1% - 17.7%) (17, 33). Other studies conducted in various parts of the world have shown that ermA and ermC were responsible for the majority of resistance to erythromycin among S. aureus isolates (4, 34, 35). In some studies, ermA is predominant, while in the others ermC is more prevalent than ermA genes (36, 37). In contrast to our finding, in all the above-mentioned studies, the rate of ermA gene was more than the rate of ermC gene. However, in agreement with our finding, a study carried out in Denmark showed 16% and 84% of S. aureus isolates were harboring ermA and ermC, respectively (37). Spiliopoulou et al. (37) have reported in Greece that ermC gene with 70% prevalence is the predominant genetic determinant compared to ermA gene with 22%. This predominance is probably due to the spread of distinctive clones (which carry ermC gene) in the mentioned countries and our region.
Based on the findings of our study and some other studies, no ermB gene has been found in studied isolates of S. aureus (17, 33, 38). But in 3 studies conducted in France, Brazil, and Turkey, the frequency of ermB gene was 0.7%, 2.2%, and 8.3%, respectively (16, 39, 40). Our finding did not show any msrA gene that is similar to the two available studies from Iran (17, 33). However, different low rates of msrA gene have been reported by other researchers (16, 38, 40).
A notable finding of the present study was the co-presence of ermA and ermC in a significant number (39, 18.14%) of our isolates. In other studies, the presence of both genes has been reported in their studied isolates with different rates (16, 17, 33, 38). Six S. aureus isolates which had iMLSB phenotype did not carry any of ermA and ermC genes, therefore, other genes or factors may have a significant role in resistance to erythromycin. Similar finding has been reported in other studies from Iran and Turkey (16, 17, 33).
5.1. Conclusions
The rate of inducible resistance to clindamycin in S. aureus isolates is relatively high in the north west of Iran. Since isolates with inducible resistance may mutate and change to constitutive resistance, for excluding inducible clindamycin resistance, microbiology laboratories must correctly recognize clindamycin susceptibility in S. aureus isolates by using D-test. ICR frequency was higher in MSSA than MRSA isolates. Our finding showed ermC as the predominant genetic determinant. This predominance is probably due to the spread of distinctive clones (which carry ermC gene) in our region.