1. Background
In the last decade, tuberculosis (TB) has recurred as one of the leading causes of death with more than two million fatalities and an annual occurrence of 9.2 million new cases (1). Various reasons such as lack of information on Mycobacterium tuberculosis as well as its widespread resistance and molecular epidemiology have led to its persistence and the subsequent spread of tuberculosis (2, 3). By specific amplification of the internal transcribed spacer (its) gene that harbors sufficient sequence diversity to distinguish most Mycobacterium species and the regions of difference (rd) gene that is only present in the M. tuberculosis complex, the identification was confirmed (4, 5).
Ethambutol (commonly abbreviated EMB or simply E) is the primarily used medication for the treatment of tuberculosis. It has been recommended as an alternative to streptomycin as a fourth drug during the initial stages of therapy until susceptibilities are known, except in the case of patients who have no individual risk factors for drug resistance and live in areas where the primary isoniazid resistance is below 4% (6, 7). Genes belonging to the emb (ABC) operon play a role in resistance against ethambutol. Among the resistant isolates, mutations in codon306 in the embB operon had a high frequency of incident (8, 9). The frequency and type of mutations leading to the resistance of M. tuberculosis help us to understand the molecular epidemiology of TB in different geographical areas.
There are many molecular techniques for the identification of sensitivity to antibiotics and the investigation of the mutations leading to resistance in bacteria. Most of the molecular methods used for the detection of genomic mutations indicative of drug resistance are based on the multiplication of a specific point of the bacterium genome followed by the analysis of the product. Polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) and direct sequencing are two of these methods (10-12).
2. Objectives
From various region of the world, different types of mutations were identified in EMB-resistant (EMBr) M. tuberculosis clinical isolates. Most of these were point mutations, and a few isolates had a deletion or insertion. In this study, we investigated the effectiveness of the PCR-SSCP method to detect mutations in the emb gene of EMBr and EMB-susceptible (EMBs) M. tuberculosis strains isolated in Isfahan. Moreover, the frequencies of mutations in their EMB gene were characterized by direct sequencing to determine the codons most frequently involved in resistance to EMB in Iran.
3. Methods
3.1. Conventional Screening Tests
Thirty-two M. tuberculosis isolates from different regions of Isfahan province were obtained from Isfahan tuberculosis center. Isolates were subcultured on Löwenstein-Jensen (LJ) media (Merck, Germany), incubated for 21 days at 37°C and specified on the basis of conventional options, i.e., staining, colony characteristics, pigmentation, growth temperature, and time of growth.
The sensitivity and resistance of M. tuberculosis isolates at concentrations of 2, 5, and 10 μg/mL of EMB (Sigma-Aldrich, Germany) were determined via the proportion method (13). All isolates were cultured in 7H9 broth medium for 10 days. Subsequently, portions of the LJ medium without and with EMB were inoculated with 0.1 mL of 10-2 and 10-4 dilutions of McFarland 1.0 Standard 3 for each isolate. The inoculated plates were then incubated at 37°C. The inhibition percentage of colony-forming units (CFU) was determined after 3 to 6 weeks of inoculation (14, 15). M. tuberculosis H37Rv (ATCC 27294) was used as the control in this study.
3.2. Preparation of Genomic DNA
The chromosomal DNA of the isolates was extracted via the “DNA mini-prep” procedure using cetyltrimethyl ammonium bromide (CTAB) as previously described and a High Pure PCR Template Preparation Kit (Roche Applied Science, Germany). A loopful of Mycobacteria on LJ media was suspended in 500 μL of sterile double-distilled water and inactivated by boiling at 80°C for 30 minutes. Five microliters of lysozyme (10 μg/mL in 10mM Tris-HCl, pH 8.0) was added to the suspension and followed by incubation at 37°C for 15 minutes. The remaining steps of purification were performed according to the manufacturer’s instructions. Under the high-salt conditions, CTAB binds the polysaccharides, removing them from the solution (16). The concentration and purity of the extracted DNA were determined using a UV-photometer (Biometra, Germany) at 260 and 280 nm. The purified DNA was stored at -70°C. The DNA of the drug-susceptible standard strain, M. tuberculosis H37Rv (ATCC 27294), was used as the control sample.
3.3. PCR of Internal Transcribed Spacer (its) Gene
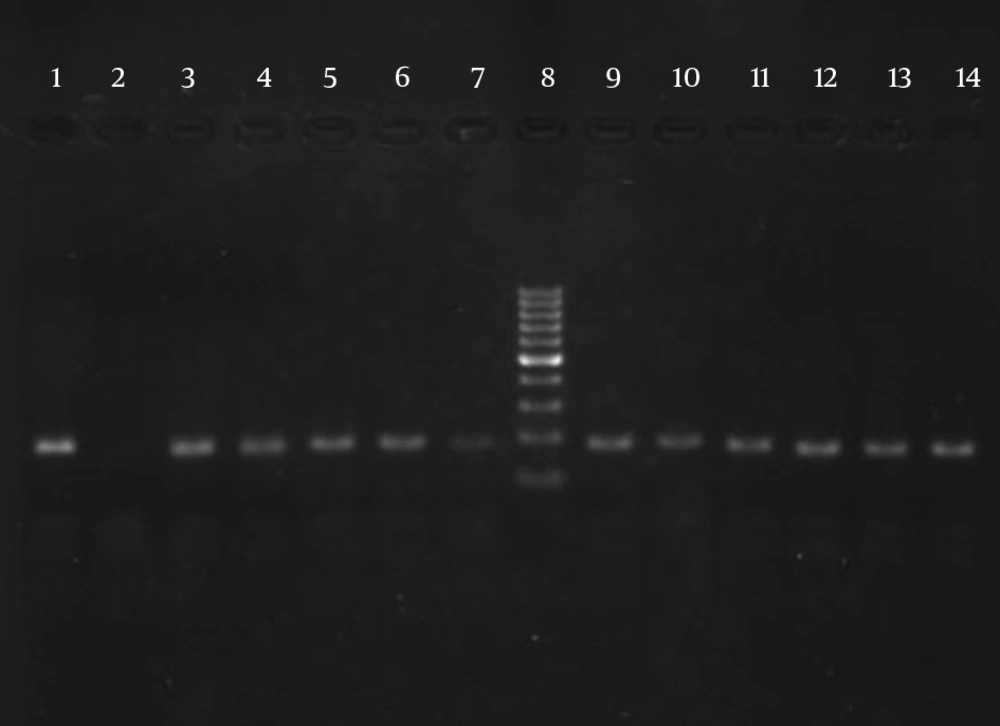
The ITS gene was amplified (185 bp) to confirm the dependency of the isolates on the M. tuberculosis complex. The targeted ITS gene region was amplified using (ITS F: CCGTGAGGGGTTCTTGTC) and (ITS R: GGCAGCGTATCCATTGATG) primer sets (Faza Biotech Company, Iran). The PCR was performed in a 50-μL reaction mixture containing 1.0 μL of each primer (10 pmol), 25 μL of master mix, 5 μL of purified DNA (20 ng), and 18 μL of RNase-free water. The amplification reactions consisted of one denaturation cycle at 94°C for 5 minutes followed by 40 cycles in the given order: 30 s at 95°C, 40 seconds at 60°C, 1 minute at 72°C, and 5 minutes at 72°C as the final extension (5). The amplicon size was analyzed by agarose gel electrophoresis (2.0%) and gel staining using ethidium bromide and UV detection.
3.4. PCR of Regions of Difference (rd) Gene
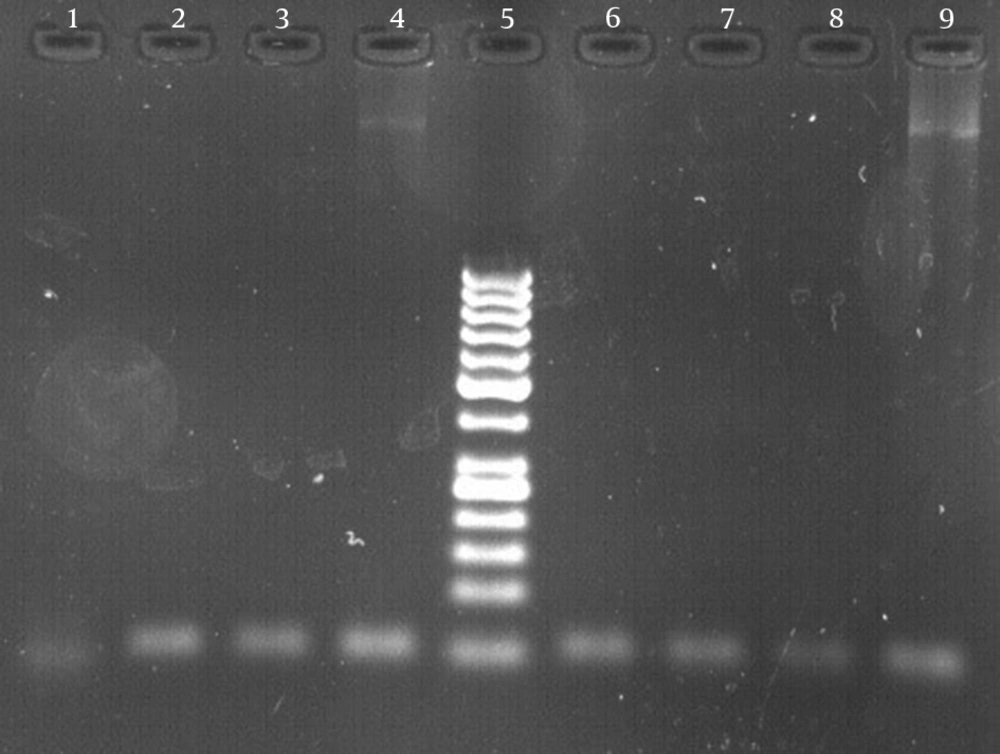
In order to determine the M. tuberculosis species among the members of the M. tuberculosis complex, a 51-bp fragment of the rd gene was amplified using rd 9 forward primer (5-4TTTCGAGCCGTAAATTACTGTG) and rd reverse primer (5-GAGCATTCTCGCTCCGAAT) (Faza Biotech Company, Iran) as the basis for ITS gene amplification (4).
3.5. EMB Susceptibility Testing of M. tuberculosis
Several colonies of all isolates were harvested from the LJ medium and suspended in phosphate-buffered saline (PBS) and homogenized thoroughly by shaking in tubes containing small glass fragments. A McFarland 1.0 concentration for the suspension of each isolate was prepared. Two 100-μL measures consisting of 10-2 and 10-4 diluted suspension were inoculated on the LJ medium supplemented with 10% oleic acid-albumin-dextrose catalase (OADC). A serial three-fold concentration of EMB (2, 5, and 10 μg/mL) and an antibiotic-free medium were prepared as the control items, inoculated by a diluted suspension of M. tuberculosis isolates, and incubated at 37°C for 3 - 6 weeks. The lowest concentration of EMB that inhibited more than 99% of the M. tuberculosis isolates was considered as the minimal inhibitory concentration (MIC).
3.6. PCR-SSCP and Direct Sequencing of embB Gene
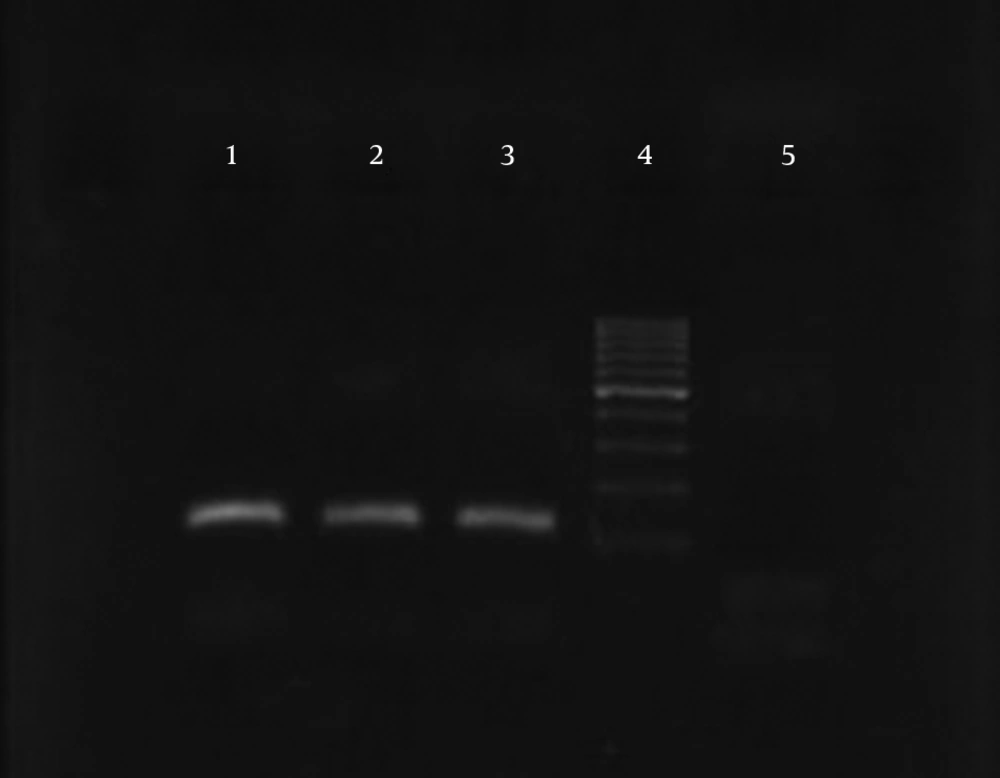
PCR-SSCP of the embB gene was performed to detect any mutation in the sequences of the resistant isolates. The sequences of the embB locus primers were EmbF (5-ATTCGGCTTCCTGCTCTGG-3) and EmbR (5-GAACCAGCGGAAATAGTTGG-3) (Bioneer Corp., Korea). The PCR reactions (50 μL) contained 10 ng of target DNA, 15 pmol of primers, 2 mM dNTP (Pharmacia Biotech), 2.5 U Taq polymerase (MBI Fermentas), 1.5 mM MgCl2, and 5 μL of 10x buffer. The reaction took place in a Hybaid thermal cycler (Omni Gene TR3SM2). The PCR was performed using initial denaturation at 95°C for 4 minutes, 45 amplification cycles for 60 seconds at 95°C, 40 seconds at 60°C and 20 seconds at 72°C, and finally, the last extension occurred at 72°C for 2 minutes (17). The PCR products were analyzed by electrophoresis on 10% (w/v) acrylamide gel via the SSCP method. The SSCP gel was prepared by mixing 10-mL 40% (w/v) acrylamide solution, 25.6-mL H2O, 4-mL 10X TBE, 30-mL TEMED, and 300-mL ammonium persulfate. Six microliters of the amplified products was mixed with 4 mL of loading buffer and loaded on the acrylamide gel at 1.2 W for 14 - 16 hours at a temperature of 4°C after which the gel was silver stained.
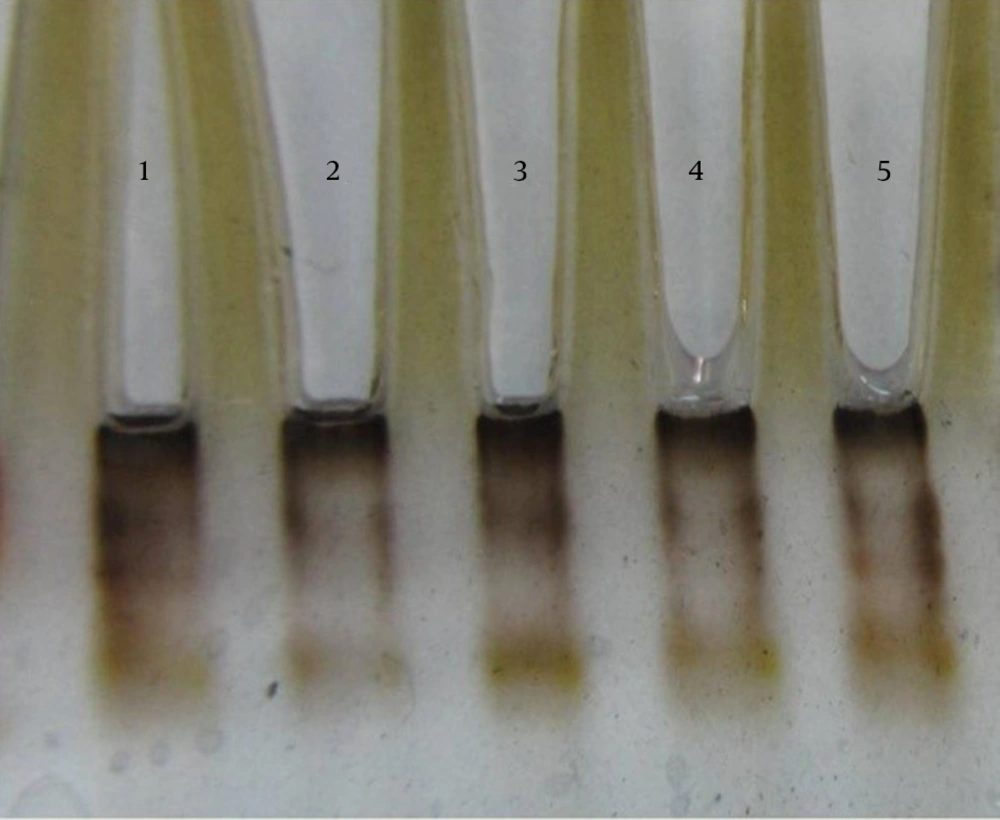
After the SSCP analysis, the PCR products of two EMBr strains and four sensitive strains were sequenced using upstream primer Emb with Applied Bio systems 377 automated sequencing protocol (Bioneer, Korea). All the postrun analyses were performed using Mega 5 and ClustalW (version 1.82), and BLASTn algorithm (http://blast.ncbi.nlm.nih.gov/). In all the stages of the experiment, M. tuberculosis strain H37RV (ATCC 27294) functioned as the control. Each sequence was compared with both the control strain sequence and published embB gene sequences (http://www.ncbi.nlm.nih.gov/genbank/).
4. Results
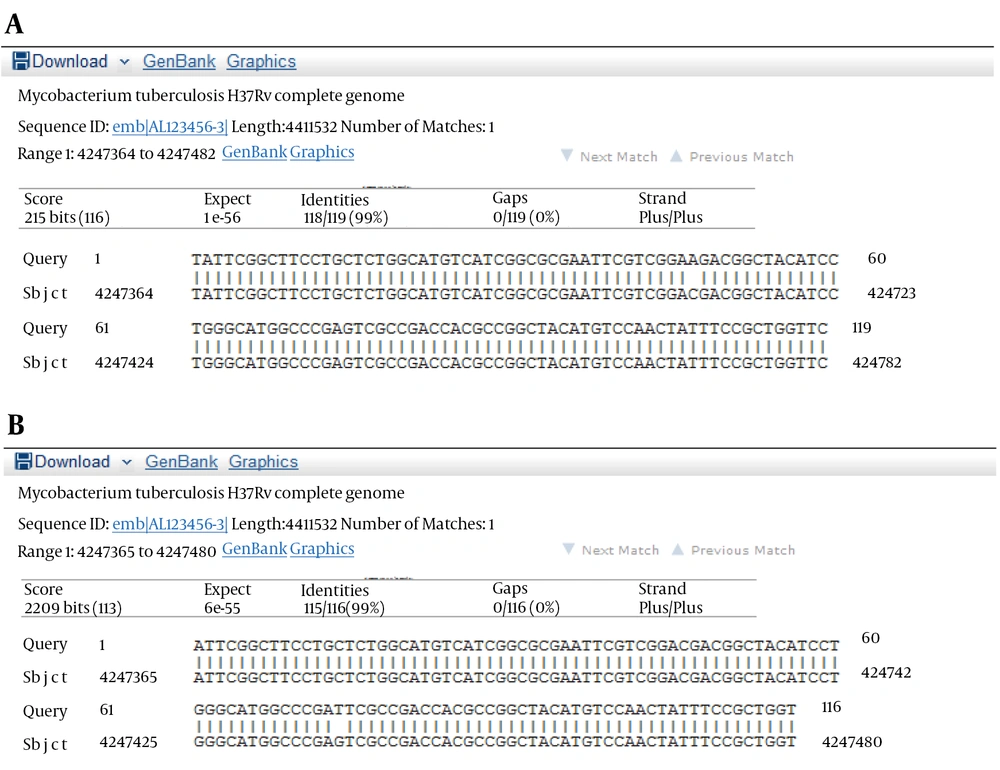
The results of susceptibility testing indicated that two isolates (6.25%) were resistant to EMB in each of the 2, 5, and 10 μg/mL concentrations of EMB. In our study, the PCR of its and rd genes was the primary technique used for the identification of M. tuberculosis isolates. The measurements of the PCR products size were 185 bp (Figure 1) and 51 bp (Figure 2) for the its and rd genes, respectively. Emb (ABC) plays a role in resistance against EMB. The PCR products of the 118-bp embB gene of the M. tuberculosis strains and their PCR-SSCP patterns are shown in Figures 3 and 4, respectively. Based on our SSCP results, two groups of bands were observed: double bands in the standard resistant strain and resistant isolates, and triple bands in the EMBs isolates of M. tuberculosis. After the SSCP analysis, two PCR products from the EMBr isolates and four from the EMBs isolates were sequenced. The nucleotide blast of the PCR products is shown in Figure 5. The sequencing results indicated mutations in two embB genes of the resistant isolates. These mutations had occurred in codons 299 and 309 (Figure 5); however, no mutation was observed in the four EMBs isolates.
5. Discussion
Mutations in the M. tuberculosis embB locus are associated with EMB resistance and the examination of sequence variation in this operon in genetically distinct susceptible and resistant organisms (18). Considering the increasing number of drug-resistant isolates of M. tuberculosis in many countries of the world and the expansion of tuberculosis cases, this study was designed to determine if polymorphisms were uniquely present in EMBr versus EMBs M. tuberculosis isolates in Iran and additionally highlight the different mutation types that lead to EMB resistance in order to specify the molecular epidemiology of tuberculosis in this geographical area.
The study was performed using a sample of 32 M. tuberculosis isolates obtained from Isfahan tuberculosis center, Iran. The isolates were initially tested for EMB susceptibility using the proportion method. The critical concentrations were 2, 5, and 10 μg/mL. Each series of tests included a control strain of M. tuberculosis: strain H37Rv (ATCC 27294). Two isolates (6.25%) were EMB resistant to all different concentrations of EMB, and 30 isolates (93.75%) were EMB susceptible. Our results are similar to those of Asgarzade et al. (19) and Namaei et al. (20), but are not in agreement with those of Bahrami et al. (21) and Xin Shen et al. (22). The findings of the survey by Tavanaee et al. (23) suggest that only 3% of the patients are EMB resistant in the east of Iran. The prevalence in our work is significantly lower than that reported in Latvia, Thailand, Mozambique, and Uganda by WHO (3) and the study by Plinke et al. (24) that obtained a resistant rate of 63.5%. The frequency of resistance was reported as 18.8% by Hazbon et al. (25).
We performed the PCR-SSCP analysis with selected EMBr clinical isolates. We showed that every two EMBr clinical M. tuberculosis isolates had mutations in the embB gene. This indicates that the embB mutation is the major mechanism of EMB resistance in M. tuberculosis, and this finding is consistent with some previous observations (8, 13). The resistant M. tuberculosis isolates showed two bands, but all 30 EMBs isolates showed 3 bands. The results of this study indicate that mutations in EMBr strains of M. tuberculosis may be readily detected by this technique. After the sequencing of 118 bp of the embB gene region of the isolates, one EMBr strain showed a single mutation in codon 299 and another strain showed a codon 309 mutations, but no mutation was observed in the sensitive isolates. Ankita Garge et al. (15) showed a good relationship (81%) between SSCP and the sequencing method. Amita Jain et al. (26) showed motion changes in isolates as compared to motion changes in the EMB sensitive standard strain (H37RV).
The results of some studies show that the region of the embB containing residue Met 306 is highly conserved in some species of Mycobacteria, including M. tuberculosis. Mokrousov et al. (27) reported a high frequency of mutation in embB codon 306 in 48.3% of the resistant strains and 31.2% of the sensitive strains. Lee et al. (28) reported no mutation in the embB gene of the susceptible isolates. Sreevatsan et al. (18) reported a point mutation in codon 306. Shen et al. (22) reported mutation in codon 306 in both the EMBr and EMBs strains of M. tuberculosis clinical isolates collected in Shanghai, China. Hazbon et al. (25) reported that mutations in embB codon 306 cannot be uniquely associated with any particular type of drug resistance. Plinke et al. (24) show that with regard to the frequency of mutation in different codons, 68% of the mutations are in codon 306. Dawei Shi et al. (29) reported that a significant number of the mutations in codons 406 and 497 occurred in the EMBr isolates without the occurrence of mutation in codon 306. They concluded that mutation in codons 406 and 497 are the signifiers of resistance to EMB. However, in our study, the selected part of the embB gene did not include codons 406 and 497. Although various studies do not agree on the role of codon 306 of the embB gene as a marker for EMB resistance, in most studies on EMBr isolates, it is found that mutation occurs in this codon.
We claim that the PCR-SSCP technique can separate resistant isolates from sensitive isolates. Sequencing results in this study showed mutation in codons 309 and 299 of the embB gene. In none of the resistant isolates, mutation was observed in codon 306. Research differences reflect the complex interaction between the drug and the target at a molecular level, where the position of the affected allele seems to be critical. Further studies are required to determine other point mutations in EMBr M. tuberculosis isolates in Iran.