1. Background
Escherichia coli is a commensal bacterium living in human and animal intestine, but is also found in many environments such as water, soil, or plants. Moreover, some E. coli strains are able to cause disease of the gastrointestinal, urinary, or central nervous system in the hosts. Intestinal pathogenic E. coli strains are divided into 6 categories as follow: Enteropathogenic E. coli (EPEC); enterohaemorrhagic E. coli (EHEC); enterotoxigenic E. coli (ETEC); enteroinvasive E. coli (EIEC); enteraggregative E. coli (EAEC); and diffusely adherent E. coli (DAEC) (1).
Among the various extraintestinal infections caused by this bacterium, urinary infections are very important in humans (2). Recent reports have indicated that the rate of antibiotic resistance among E. coli isolates is increasing rapidly (3). The main reason for this problem is the widespread clinical use of antimicrobials, which leads to the suppression of susceptible normal flora and rise of resistant strains (4).
Tetracyclines are broad- spectrum antibiotics that act through inhibition of protein synthesis. The suitable antibacterial characteristics of these antibiotics and the absence of any important side effects have led to their widespread use in treating human infections (5). Efflux pumps are transmembrane proteins, which export toxic materials such as antibiotics out of the bacterial cell. Among the prokaryotes, 5 major efflux pumps are detected as follow: MFS (major facilitator super family); MATE (multidrug and toxic efflux); RND (resistance-nodulation-division); SMR (small multi drug resistance); and ABC (ATP binding cassette) (6). Efflux pumps are the most abundant in Gram-negative bacteria, but ribosomal protection mechanisms of resistance is more prevalent in Gram-positive bacteria (7).
Tetracycline efflux pumps belong to FMS group, and most of them export tetracycline from the bacteria but not to minocycline. Only the product of the tetB gene could efflux both antibiotics out of the cell (5). These pumps belong to 6 groups according to amino acid sequence similarities: tetA, tetB, tetC, tetD, tetE, tetG, tetH, tetJ, tetZ, tetI, and tet30 are placed in group 1 (5).
2. Objectives
In the present study, the occurrence of phenotypic tetracycline resistance and carriage of tet resistance genes in uropathogenic E. coli strains was investigated in Iranian patients.
3. Methods
3.1. Isolation and Identification of Uropathogenic E. coli
A total of 50 uropathogenic E. coli isolates from human urinary samples were obtained from the microbiology laboratory of Azerbaijan hospital, Urmia, Iran, from October 2014 to February 2015. All samples belonged to hospitalized patients aged 20 to 40 years. The urine samples were streaked on to MacConkey agar and incubated at 37°C for 24 hours. Then, the plates were examined for E. coli red colonies with a dark red center. Other biochemical tests like indol, methyl red, voges-proskauer, Simmons citrate, and TSI were conducted to confirm the presence of E. coli (8).
3.2. Molecular Characterization of Uropathogenic E. coli
Escherichia coli confirmation was achieved by PCR assay (9). E. coli cultures were grown overnight at 37°C, and the chromosomal DNA was extracted using bacterial genomic purification kit (iNtRON, Korea). Then, the extracted DNA was used as a template for PCR amplification of uropathogenic E. coli 16s rRNA gene by forward primer F: 5/- GTA TAG ATA CCC TGG TAG TCCA-3,/ and reverse primer R: 5/- CCC GGG AAC GTA TTC ACC G-3/. PCR reactions were carried out in the total volume of 25 µL using DNA thermo cycler (MWG AG BIOTECHTHERMAL CYCLER, USA).The conditions used for the PCR assay are as follow: 3 minutes of initial denaturation at 95°C, followed by 26 cycles of 94°C for 1 minute; 55°C for 1 minute, and 72°C for 10 minutes (9). The PCR products were separated by 1% agarose gel electrophoresis stained with red safe (Sigma Aldrich, Germany) and visualized under UV exposure.
3.3. Antibiotic Susceptibility Testing
Tetracycline susceptibility profiles of isolated uropathogenic E. coli were determined via the disc diffusion method (10). A 15 mm Muller-Hinton medium plate was swabbed with nutrient broth inoculated with E. coli and incubated to a turbidity of 0/5 McFarland standard. Commercially tetracycline disks (30 µg) were placed on the inoculated plates (Padtan Teb, Iran). Then, all the plates were incubated aerobically at 37°C for 18 - 20 hours. The diameters zone of inhibition (mm) around the disks were measured and interpreted by referring to the performance standard for antimicrobial susceptibility testing, as described by the clinical and laboratory standards Institute (CLSI) guidelines, 2015 (11). The percentage of E. coli resistant, intermediate and susceptible to tetracycline was determined.
3.4. Identification of Tetracycline Efflux Genes using PCR Assay
3 PCR amplification assays were performed to determine the prevalence of tetA, tetB, tetD, tetE, tetH, tetG, and tetZ among all tetracycline resistant E. coli. Table 1 demonstrates the sets of primers used to identify these determinants (12). The first PCR assay performed to amplify the tetB and tetE genes is as follow: Initial denaturation at 94°C for 1 minute, 25 cycles each consisting of minute at 94°C, minute at 50°C, and 72°C for 10 minutes. The second PCR used to detect tetA and tetD genes was performed as above, but the annealing temperature was 48°C.
| Primer | Gene | Sequences |
|---|---|---|
| Forward | tetA | 5/ GCGCGATCTGGTTCACTCG 3/ |
| Reverse | tetA | 5/ AGTCGACAGYRGCGCCGGC 3/ |
| Forward | tetB | 5/ TACGTGAATTTATTGCTTCGG 3/ |
| Reverse | tetB | 5/ ATACAGCATCCAAAGCGCAC 3/ |
| Forward | tetC | 5/ GCGGGATATCGTCCATTCCG 3/ |
| Reverse | tetC | 5/ GCGTAGAGGATCCACAGGACG 3/ |
| Forward | tetD | 5/ GGAATATCTCCCGGAAGCGG 3/ |
| Reverse | tetD | 5/ CACATTGGACAGTGCCAGCAG 3/ |
| Forward | tetE | 5/ GTTATTACGGGAGTTTGTTGG 3/ |
| Reverse | tetE | 5/ AATACAACACCCACACTACGC 3/ |
| Forward | tetG | 5/ GCAGAGCAGGTCGCTGG 3/ |
| Reverse | tetG | 5/ CCYGCAAGAGAAGCAGAAG 3/ |
| Forward | tetH | 5/ CAGTGAAAATTCACTGGCAAC 3/ |
| Reverse | tetH | 5/ ATCCAAAGTGTGGTTGAGAAT 3/ |
| Forward | tetJ | 5/ CGAAAACAGACTCGCCAATC 3/ |
| Reverse | tetJ | 5/ TCCATAATGAGGTGGGGC 3/ |
| Forward | tetZ | 5/ CCTTCTCGACCAGGTCGG 3/ |
| Reverse | tetZ | 5/ ACCCACAGCGTGTCCGTC 3/ |
The third PCR regimen used to amplify tetH, tetG, and tetZ was carried out as described above, but the annealing temperature was 46°C. Finally, the reaction products were run on 1% agarose gel, and the prevalence of each tetracycline efflux gene was determined using agarose gel electrophoresis. Descriptive statistics, such as the percentage of tetracycline efflux genes, were done using the statistical package, SPSS, Version 15.0.
4. Results
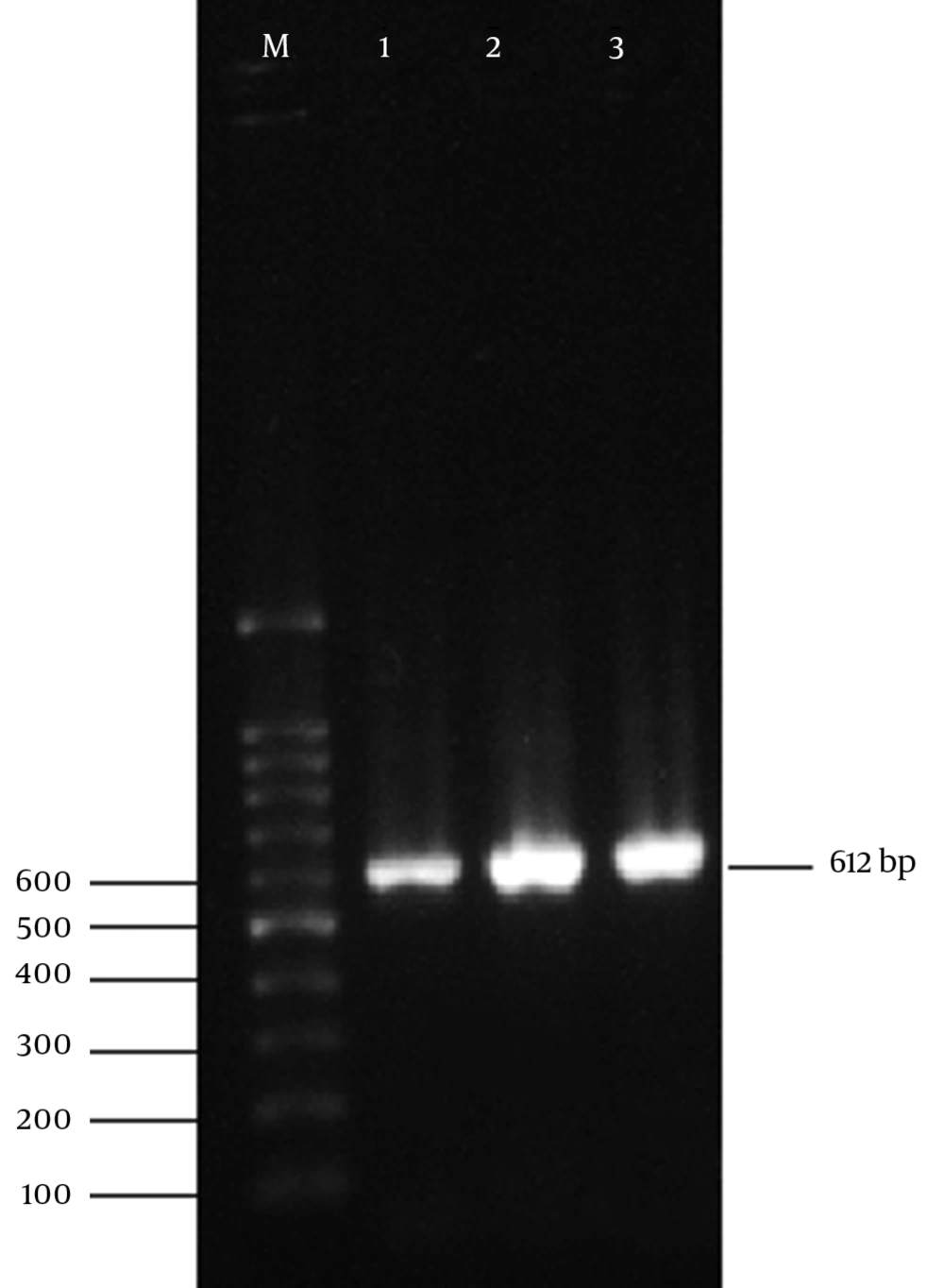
All 50 isolated bacteria from urinary infections, possessed the cultural, morphological, and biochemical characteristics of E. coli. Also, 16s rRNA gene of E. coli were detected in all of the isolates by PCR assay using 16s rRNA universal primers. Figure 1 displays the product of 16s rRNA gene found at 612 bp using 100 bp DNA marker.
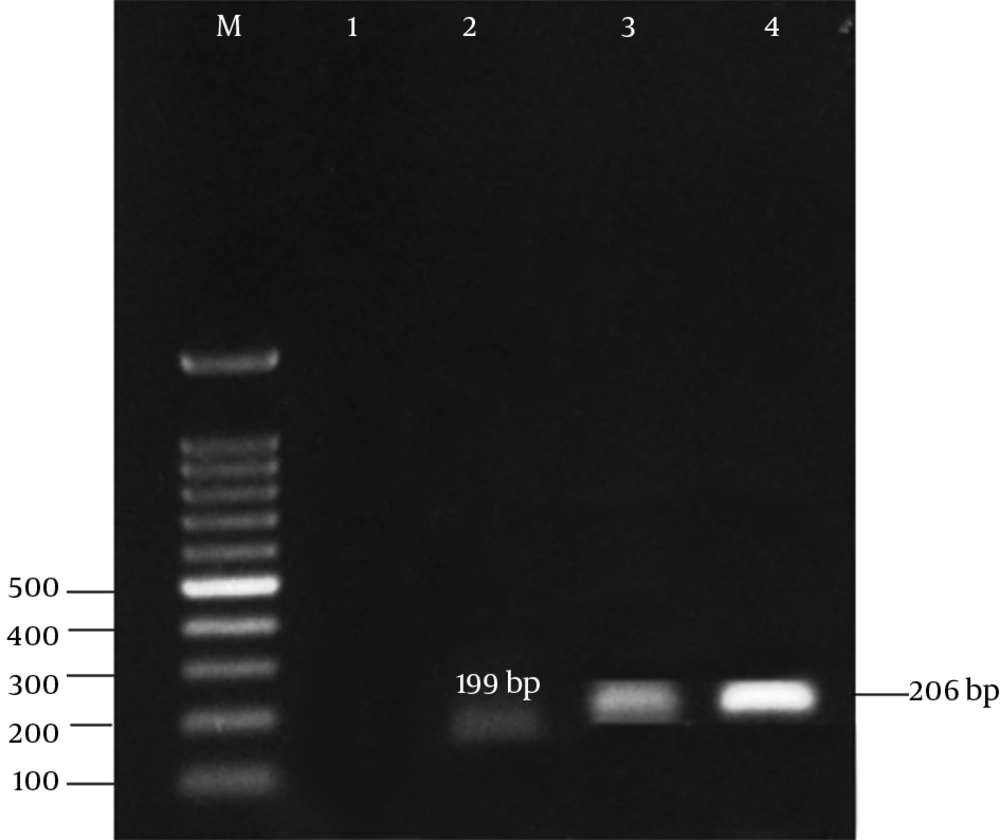
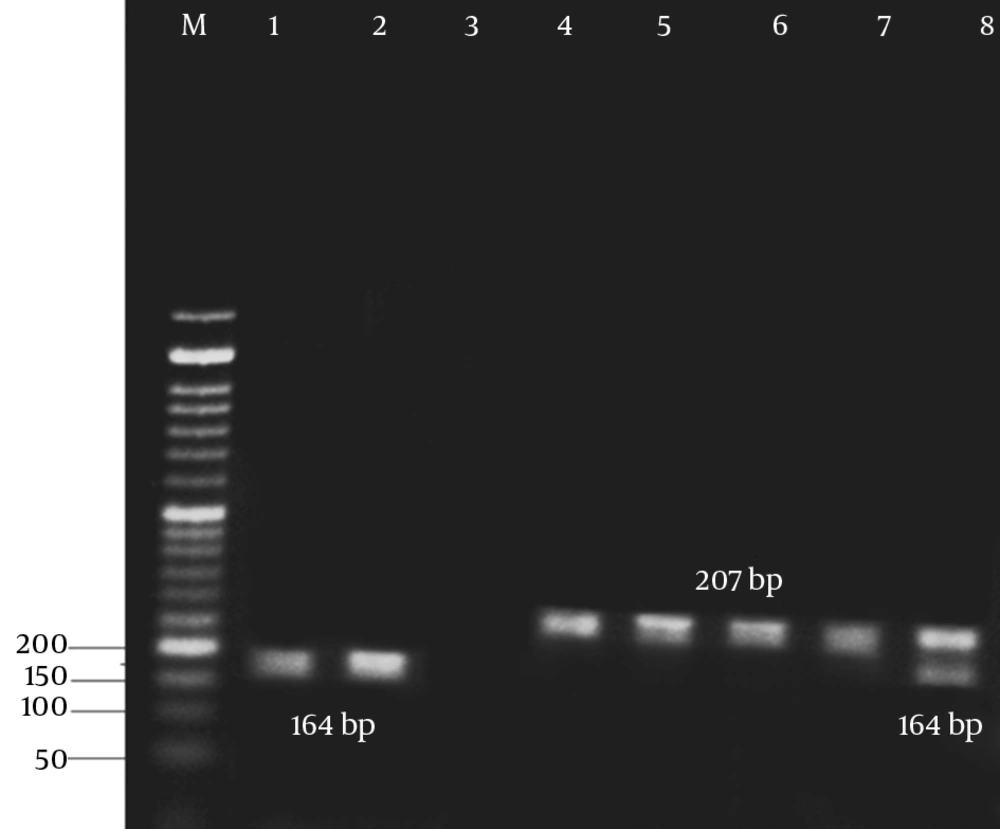
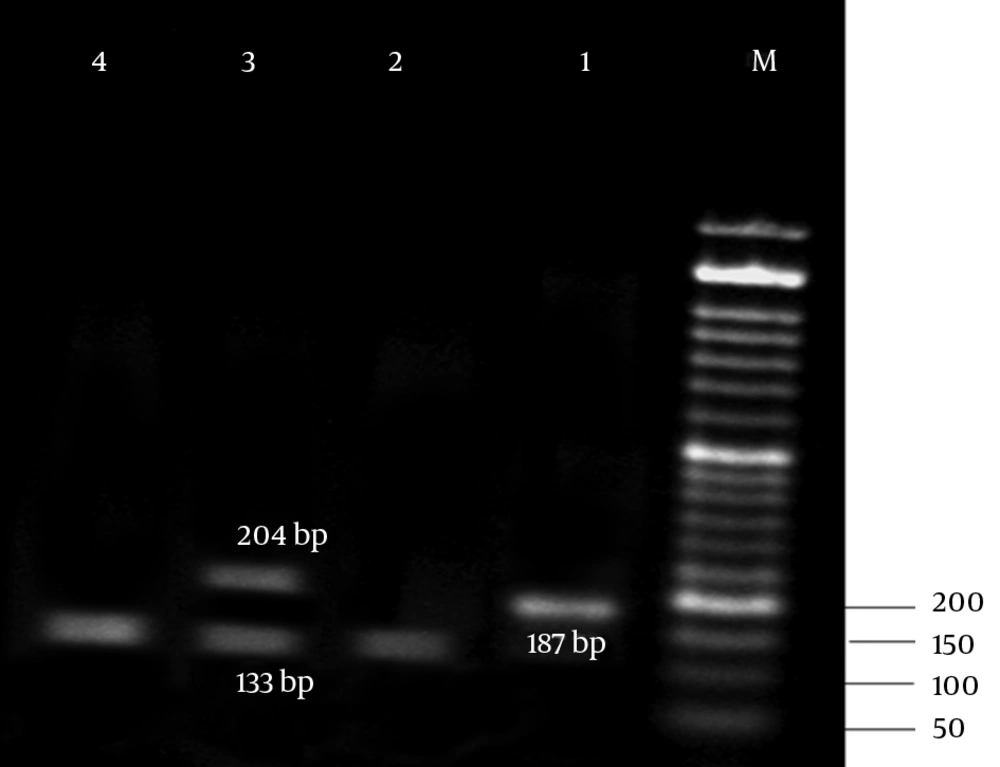
The results of antibiotic susceptibility testing in E. coli isolates showed that 31 (62%) of them were phenotypically resistant to tetracycline, 16 (32%) were susceptible, and 3 (6%) showed intermediate susceptibility to tetracycline. Three multiplex PCR assay were developed to monitor the prevalence of resistance determinants for tetracycline. The results are presented in Figures 2 - 4.
The frequency and distribution of tetracycline resistance genes among E. coli strains used in the presented study are demonstrated in Table 2.
| Sample | tetZ | tetG | tetH | tetD | tetC | tetA | tetE | tetB |
|---|---|---|---|---|---|---|---|---|
| Frequencya | 14 | 16 | 8 | 11 | 12 | 32 | 12 | 36 |
aValues are expressed as %.
The results of the present study revealed that all tetracycline resistant isolates carried at least 1 of the tet determinants examined; tetB was the most prevalent gene detected in 36% of the resistance isolates, followed by tetA (32%), and tet A + B (18%). Moreover, tetC, tetE, tetG, tetD, and tetZ were found to have lower frequencies. However, the lowest prevalent gene among the studied determinants belonged to tetH (8%).
5. Discussion
Among 50 E. coli strains isolated from patients with urinary infections, 31 strains showed resistance to tetracycline. Monitoring the distribution prevalence of tetracycline resistance genes, we found that the tetB gene had the highest frequency among the studied determinants. Escherichia coli is a facultative Gram-negative bacterium, which can cause many infections, like diarrhea, urinary tract infections, meningitis, peritonitis, and septicemia in humans (13). However, the most prevalent form of the extraintestinal infection caused by E. coli is urinary tract infection (14).
Antibiotic resistance in E. coli has emerged and increased in many countries and led to the difficult treatment of infectious bacteria (2). In the present study, 62% of human E. coli strains were resistant to tetracycline, which supported the above hypothesis. Tetracyclines are broad-spectrum antibiotics that are used for treatment and prevention of human infections (15). Chlortetracycline was the first tetracycline identified in the late 1940s (16). However, heavy clinical use of tetracyclines has a major role in the emergence and dissemination of tetracycline resistance among infectious bacteria (17).
Different tet genes are responsible for tetracycline resistance in Gram- negative bacteria like E. coli (15). However, the most common tet resistance mechanism in E. coli is tetracycline efflux pumps, which exports the drug out of the cell (17). These proteins belong to 6 groups: tetA, tetB, tetC, tetD, tetG, tetH, tetZ , tetE, tetI, and tet30 that are placed in Group 1. Several investigations have studied tetracycline resistance in bacteria by examining their ability to grow in the presence of drugs, but these studies are not useful for determining the types of tetracycline resistance determinants responsible for antibiotic resistance in bacteria (7). On the Other hand, molecular microbiology techniques can accurately describe genetic basis of antibiotic resistance in bacteria (18).
In the present study, the molecular PCR assay was also used to determine the tetracycline resistance genes prevalence among E. coli isolates. In 2006, Karami et al. studied the prevalence of tetracycline resistance genes in intestinal E. coli strains and identified tetB and tetA as the most common tetracycline resistance genes in human E. coli (10). Our findings about the frequency of tetracycline resistance genes in urinary E. coli are in agreement with their results.
In 2007, Tuckman et al. examined the occurrence of tetracycline resistance genes among E. coli isolated from patients and found that tetA and tetB had the highest frequency among the studied determinants. In the present study, the same determinants had the highest occurrence among tetracycline efflux genes belonging to Group 1. The results of monitoring the prevalence of tetracycline resistance genes in E. coli strains isolated from human urinary infections, confirmed the emergence and spreading of the resistant E. coli in human populations.