1. Background
Members of the genus Trichophyton are the most common agents of dermatophytosis in humans and other animals, and are associated with a variety of clinical aspects (1-3). The most frequent species of dermatophytes are Trichophytonmentagrophytes, T. tonsurans, and T. rubrum, and these species cause a multiplicity of cutaneous disorders. They are keratinophilic fungi that attack keratinized tissue and cause a wide spectrum of clinical manifestations that vary from mild to severe, but infections are not life-threatening. Owing to the high degree of phenotypic similarity among species, identification is difficult. Conventional approaches for species-level identification in the diagnostic laboratory are based on morphological and physiological criteria, require several days or weeks to obtain results, and are frequently unspecific. To overcome these problems, molecular techniques have recently been developed to rapidly and precisely identify species of Trichophyton that potentially cause human infection.
Numerous recent articles and reviews have exhaustively discussed the various molecular techniques available for dermatophyte species identification. Molecular tools, such as sequencing of the internal transcribed spacer (ITS) region of ribosomal DNA (rDNA), have shown promise for species identification in all genera of dermatophytes, and is relatively expensive (4-7). Moreover, ITS-based analyses have found that sequence variation is limited to only one or a few single nucleotide polymorphisms (SNPs) between certain species, e.g., T. tonsurans and T. equinum or T. rubrum and T. soudanense (7). This limited genetic variation suggests that the development of alternative molecular tools with sufficient specificity, reproducibility, and sensitivity is highly necessary (8). Rolling-circle amplification (RCA) is a powerful and simple procedure for distinguishing closely related taxa at the species level; it is based on the rolling replication of short single-stranded DNA circles via specific DNA polymerases under isothermal conditions. This enables the detection of target nucleic acid sequences, including SNPs, with high specificity (9-12).
2. Objectives
The objective of the current study is to establish the use of RCA based on ITS rDNA to rapidly identify Trichophyton species that potentially cause human and animal disorders.
3. Materials and Methods
3.1. Fungal Strains
In the current study, all samples were obtained from different human sources that were referred to the mycology laboratory at Tooba Clinic, Mazandaran University of Medical Sciences, Iran and were suspected dermatophytosis cases. The study protocol was approved by the Medical Research Ethics Committee of Mazandaran University of Medical Sciences (Ethical No. 90-2-28/90-14) and since the laboratory diagnosis was part of the patients’ routine care, informed consent for research purposes was not specifically obtained. A total of 61 isolates belonging to three species of Trichophyton including the three reference strains T. rubrum (Centraalbureau voor Schimmelcultures: CBS 130927), T. mentagrophytes var. interdigitale (National Biological Resource Center: NBRC 5812), and T. tonsurans (NBRC 5928) were examined.
3.2. Morphological Identification
All clinical samples from suspected patients were cultured on plates of Sabouraud glucose agar (Merck, Darmstadt, Germany), with chloramphenicol and cycloheximide (SCC). The plates were incubated for 4 – 6 weeks at 28°C. All colonies were examined macroscopically and microscopically in Lactophenol Cotton Blue. The Trichophyton species were identified based on microscopic morphology and in vitro tests including urease and hair perforating tests, as required.
3.3. Molecular Identification
3.3.1. DNA Extraction
First, Trichophyton species were grown on Sabouraud dextrose agar (SDA, Difco, USA) for 10 days at 24°C in dark conditions. A sterile blade was used to scrape off the hyphae from the surface of the plate, which were transferred to a 2-mL Eppendorf tube containing 1 mL of lysis buffer (200 mM Tris-HCl, pH 8.0, with 25 mM EDTA, 0.5% [wt/vol] sodium dodecyl sulfate, and 250 mM NaCl). Cells were mechanically disrupted with a conical grinder for approximately 1 min, and then incubated at 100°C for 15 min. Next, 150 μL of 3.0 M sodium acetate buffer was added, the mixture was vortexed and incubated for 10 min at -20°C, and the solution was mixed and centrifuged for 5 min at 10,000 ×g. The supernatant was transferred to a new tube and phenol/chloroform (1:1, v/v) was used for extraction. DNA was allowed to precipitate with an equal volume of isopropanol for 10 min at -20°C and then centrifuged for 5 min at 10,000 rpm. The pellets were washed with cold 70% ethanol, dried at room temperature, resuspended in 97.5 mL of TE-buffer with 2.5 mL of RNAse (20 U/mL), and incubated for 5 min at 37°C. DNA extracts were stored at -20°C prior to use.
3.3.2. ITS rDNA Amplification
The ribosomal DNA internal transcribed spacers (i.e., the ITS region of rDNA) were amplified using the universal primers ITS1 (5’-TCC-GTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) (13). PCR reactions were performed on a TC-312 thermal cycler (Techne, Duxford, Cambridge, United Kingdom) in 25-mL volumes containing 25 ng of template DNA, 2.5 mL of reaction buffer (0.1 M Tris-HCl, pH 8.0, 0.5 M KCl, 15 mM MgCl2, 0.1% gelatin, and 1% Triton X-100), 0.2 mM of each dNTP, and 2.0 U of Taq DNA polymerase. Amplification was performed as follows: 2 min at 94°C for primary denaturation, followed by 35 cycles at 94°C (45 s), 52°C (30 s), and 72°C (120 s), with a final 7-min extension step at 72°C. PCR products were visualized by 1.5% (w/v) agarose gel electrophoresis in TBE buffer, stained with ethidium bromide (0.5 μg/mL), and photographed under UV transillumination.
3.3.3. Primers and Padlock Probes Used for RCA
The approximate length of the three specific circular oligonucleotide probes for T. rubrum, T. mentagrophytesvar. interdigitale, and T. tonsurans used in this study was 96 to 102 bp, and comprised two target-complementary segments connected by a genetic linker sequence. They were previously designed by Kong et al. (8) to minimize similarity between closely related strains and to allow primer binding during RCA. However, specific padlock probes targeting the ITS1 and ITS2 regions were modified, and RCA primers designed to specifically bind the linker region of the probes were synthesized by Anaspec Inc. (San Jose, CA, USA) (Table 1).
| Probes/Primers | Sequence |
|---|---|
| T. tonsurans | (5’p-AAGCCGGAATCGCGGCCTGGgatcatgcttcttcggtgcccattacgaggtgcggatagctaccgcgcagacacgatagtctaCCCATTCGCCTAGA-3’) |
| T. rubrum | (5’p-TTGGCTGCCCATTCGCCTAGgatcatgcttcttcggtgcccattacgaggtgcggatagctaccgcgcagacacgatagtctaTGAGGGCGCTGAA-3’) |
| T. mentagrophytes | (5’p-AGCCACTAAAGAGAGGCTCGCgatcatgcttcttcggtgcccattacgaggtgcggatagctaccgcgcagacacgatagtctaCGGTCCAGCGTTT-3’) |
| RCA1 | (5’-ATGGGCACCGAAGAAGCA-3’) |
| RCA2 | (5’-CGCGCAGACACGATA-3’) |
3.3.4. Ligation of Padlock Probes
Purified PCR products (1 µL) were mixed with 2 U Pfu (1 µL) DNA ligase (Bioneer ISO 13485, Alameda, CA, USA) and 1 µL of padlock probe in 500 mM Tris-HCl (pH 7.5), 50 mM KCl, 100 mM MgCl2, 2.5 µL/mL bovine serum albumin (pH 7.5), 10 mM ATP, and 50 mM DTT with a total reaction volume of 10 μL. Multiple cycle ligations were conducted with one cycle of denaturation at 94°C of 5 min, followed by five cycles at 94°C for 30 s and 4 min of ligation at 62°C.
3.3.5. Hyper-Branched or Rolling Circle Amplification Reactions
RCA reactions were performed in a 25-μL volume containing 8 U of Bst DNA Polymerase (New England BioLabs, Ipswich, MA, USA), 200 µM deoxynucleoside triphosphate mix, 1 µL of each RCA primer, and 2 µL of ligation product. Probe signals were amplified by incubation at 65°C for 60 min and 85°C for 2 min, and the accumulation of double-stranded DNA products was detected by electrophoresis on 1% agarose containing ethidium bromide (Sigma, St. Louis, MO, USA). Ladder-like patterns were interpreted as positive reactions, while negative reactions showed no illumination.
4. Results
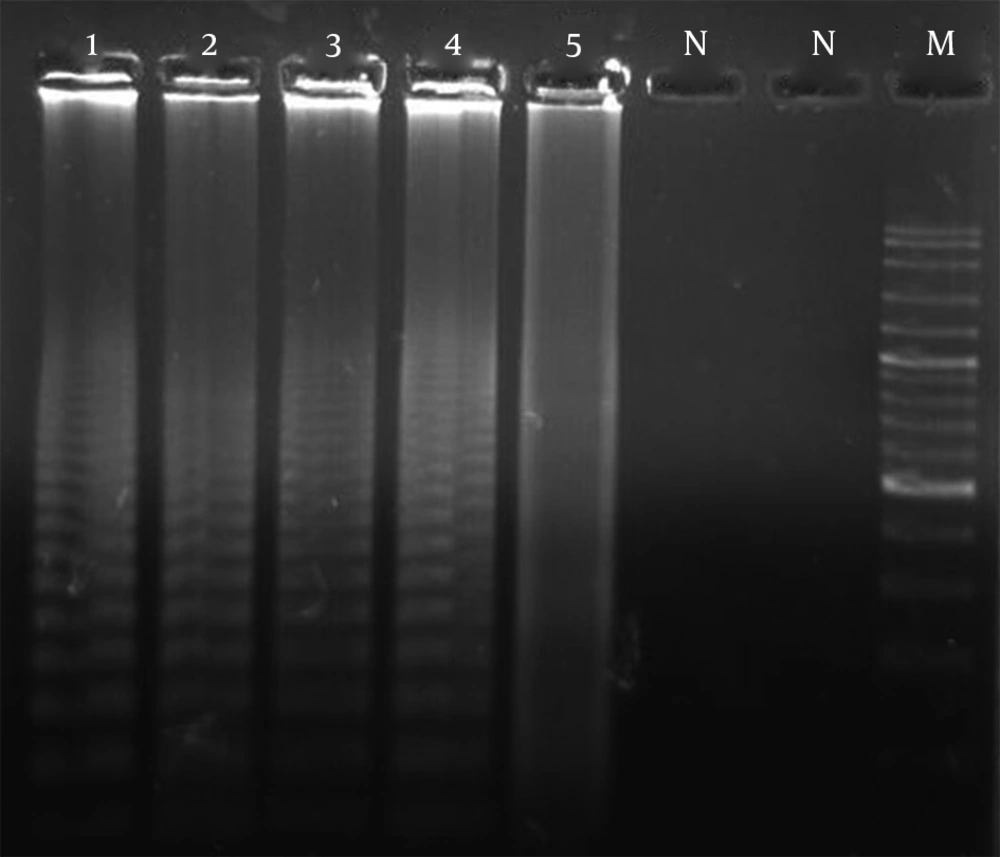
The Trichophyton species were not easily differentiated from each other based on mycological criteria. Of the 61 clinically isolated samples, 31 isolates (50.8%) were identified as T. mentagrophytes var interdigitale, 11 isolates (18.2%) as T. rubrum, 9 isolates (14.7%) as T. tonsurans, and 1 isolate as (1.8%) T. violaceum. Moreover, 9 isolates (14.7%) were identified as non-Trichophyton species. RCA and subsequent detection by gel electrophoresis yielded positive results, and the test proved to be 100% specific for each species. Neither cross-reaction between the examined species of Trichophyton nor false positive or false negative results were observed (Figure 1). Positive RCA reactions were visualized by UV irradiation as ladder-like, strongly illuminated smears on 1.2% agarose gels, while the background remained clean for negative reactions (Figure 1). RCA reactions were performed for all isolates; 45 isolates (73.3%) were positive for the probe specific to T. mentagrophytes, 8 isolates (13.1%) for the probe specific to T. rubrum, and 4 isolates (6.6%) for the probe specific to T. tonsurans (Figure 1). However, four isolates (8.6%) did not test positive using any of the three species-specific padlock probes and probably represented separate dermatophyte species.
Representative Gel of Specificity of RCA Amplification Probes. 1) Positive reaction standard for T. mentagrophytes var. interdigitale (NBRC5812) with the T. mentagrophytes var. interdigitale probe; 2) Positive reaction standard for T. rubrum (CBS130927) with the T. rubrum probe; 3) Positive reaction standard for T. tonsurans (NBRC5928) with the T. tonsurans probe; 4) Positive reaction for an unknown case with the T. mentagrophytes var. interdigitale probe; 5) Positive reaction for an unknown case with the T. rubrum probe; N) Negative reaction standard for T. tonsurans (NBRC5928) with the T. rubrum probe; N) Negative reaction standard for T. rubrum (CS 82) with the T. mentagrophytes var. interdigitale probe; M) Ladder (100 bp).
5. Discussion
Members of the genus Trichophyton are the most common agents of dermatophytosis in humans and other animals. Identification of Trichophyton species is essentially based on macroscopic and microscopic observations of their morphological features; however, the identification is complicated and laborious owing to the morphological similarity, polymorphism and cultural variability of Trichophyton species. Thus, accurate identification is time-consuming and requires a significant level of knowledge and technological expertise (4). ITS-based analyses have found that sequence variation is limited to only one or a few SNPs between certain species, such as between T. tonsurans and T. equinum, and between T. rubrum and T. soudanense (7). Although sequencing of the ITS region of rDNA is currently the gold standard for the identification of Trichophyton species and relatives, the technique is relatively expensive and time-consuming, and is impractical for the analysis of large numbers of isolates for epidemiological studies. To overcome these problems, molecular biology-based techniques have been developed for rapid and accurate species determination.
Recently, isothermal techniques, such as loop-mediated amplification and RCA, have been applied for rapid identification. The use of circularizable oligonucleotides or padlock probes is based on the rolling replication of short single-stranded DNA circles by certain DNA polymerases under isothermal conditions to detect target nucleic acid sequences, including SNPs, with high specificity (9-16). Such probes comprise two sequences that are complementary to the 5’ and 3’ termini of the target sequence joined by a genetic linker region. Upon hybridization to the target, the two probe ends become juxtaposed and are joined by DNA ligase to form a closed molecule. The intensity of the signal generated by the probe is then increased exponentially by hyper branching or RCA, and 109-fold signal amplification of each circle can be achieved within 90 min (8, 13, 17). The RCA technique was initially established by Fire and Xu (18) and Liu et al. (19) and is an isothermal in vitro DNA amplification method. It is one of a series of robust and simple techniques for distinguishing closely related taxa at the species as well as the strain level. RCA is a rapid (requiring less than 1 working day), specific (to the single-nucleotide level), and economical (requiring minimal equipment) tool for fungal screening (20, 21).
Previous studies have shown that Trichophyton species have several unique nucleotide positions suitable for the development of specific padlock probes for species characterization; these can be used to distinguishing closely related species (8). In this study, species-specific padlock probes were used to distinguish three species of Trichophyton, T. mentagrophytes, T. rubrum, and T. tonsurans. Using species-specific probes, we correctly identified all clinical isolates. Three standard species of Trichophyton, T. rubrum (CBS 130927), T. mentagrophytes var. interdigitale (NBRC 5812), and T. tonsurans (NBRC 5928) were also positively identified using only the species-specific padlock probes. The sequencing results for the ITS regions of rDNA showed 100% concordance with the RCA results. Additionally, these results were perfectly concordant with phenotypic identification. The RCA procedure required less than one working day, including DNA extraction, PCR amplification, hybridization, ligation of padlock probes, and RCA amplification, rather than sequencing.
While the results of this study can be applied generally, there were some important limitations. In particular, as mentioned by Kong et al. (8), the specificity of the T. tonsurans probe (Tt-ITS2) is ambiguous because T. equinum is highly closely related. The two species differ by only a single base in the ITS1 region (7, 22); therefore, the ITS region does not have sufficient discrimination ability. Designing padlock probes that target other genes, such as β-tubulin (BT2) and translation elongation factor 1-α (TEF1), may be beneficial. In conclusion, species identification of Trichophyton is important for epidemiological and phylogenetic purposes and for genotype delineation. Despite the shortcomings of current molecular identification systems, there is a strong stimulus for the continued use of ITS polymorphisms to generate identification barcodes. Therefore, the RCA-based assay is an alternative to DNA sequencing for species identification.