1. Background
Rotaviruses are major causative agents of diarrhea, effecting human and animals, which has a high risk for the livestock industry and public health (1). Bovine rotaviruses are members of genus Rotavirus within the Reoviridae family, with a diameter of 70 - 75nm, icosahedral and non-enveloped. The genome is a double stranded RNA having 11 segments, which encodes six structural viral proteins (VP1-4, VP6, and VP7) and six non-structural proteins (NSP1-6) (2, 3). The structural viral proteins VP4, VP6 and VP7 are important as they are used for serological characterization of rotaviruses (4). In reference to the immunity development, VP4 and VP7 are helpful, so information of these genotypes is necessary to develop a vaccine (5). When replicating the rotavirus, NSP2 is an important protein (6).
Rotaviruses are categorized into seven groups, named as A to G on the basis of VP6. The groups A to C are reported as zoonotic while groups D to G are specific for animals. Two proteins, VP4 (protease sensitive) and VP7 (glycoprotein), present in outer capsids, are used for classification of group A rotaviruses into P and G genotypes, respectively (7). Genetic characterization is important to develop preventive and diagnostic measures because bovine rotaviruses have evolved by different mutation mechanisms (8). Diversity of rotaviruses is due to interspecies transmission. Advanced typing methods are needed to identify new strains of rotaviruses (9).
Group A bovine rotaviruses cause neonatal calf diarrhea, which is economically an important disease of calves causing mortality. So it is one of the leading factors of a significant economic loss. Due to neonatal calf diarrhea, annual losses of approximately $ 9.5 million USD occurred worldwide (10, 11). In neonatal calves, about 5 - 20% of the mortality rate is observed due to bovine rotaviruses in calves. The average incidence and prevalence of the infection is 30 - 40% (12, 13). Three countries in the subcontinent (India, Bangladesh and Pakistan) account for > 30% of all rotavirus-related deaths worldwide (14). The overall prevalence of bovine rotavirus infection in Pakistan is 2.6%. Higher prevalence of 3% was recorded in Okara and Rawalpindi districts, but only 2% in Lahore (15).
Pakistan is a developing country and the cattle and buffalo calves’ population in the country is devastatingly affected by the neonatal calf diarrhea due to rotavirus outbreaks. So survival of calves is really important to produce milk, meat and hides for propagation of livestock. The aim of the current study was to detect the bovine rotavirus from fecal samples of diarrheic calves by antigen capture ELISA and molecular investigations to check the prevalence and genome sequencing of bovine rotavirus. The goal is to save the life of neonate calves from the deadly rotavirus infection for better progeny and sufficiently healthy livestock. Still, no information is available regarding the sequence data and group of circulating bovine rotavirus amongst the livestock population of Pakistan. This is the first study used to identify and assign the group to bovine rotavirus via VP4 and VP6 gene sequencing in Pakistani isolates.
2. Methods
2.1. Collection and Transportation of Samples
The 200 diarrheic fecal samples (rectal swabs and fecal material) from cattle and buffalo calves were collected aseptically from public/private sector buffalo and cattle farms from 10 districts of Punjab and placed in separate sterile tubes containing the transport medium and sealed plastic bags. The samples were transferred to the quality operations Lab (QOL), UVAS Lahore, on ice packs and kept at -20°C before further processing. Processing of samples was done for screening, isolation and identification for bovine rotavirus by making 1:3 (v/v) dilution in PBS (pH = 7.2). Supernatant was collected by homogenization and clarification through centrifugation at 5000 rpm for 15 minutes. After processing, the direct sandwich ELISA was performed to detect bovine rotavirus by using commercial rotavirus detection kit (Cypress diagnostics, Hulshout, Belgium).
2.2. RNA Extraction and cDNA Synthesis
ELISA positive bovine rotavirus samples were processed for RNA extraction with TRIzol reagent (molecular research center, Inc. USA) method as described by the manufacturer. RNA concentration was estimated using nano-drop (Thermo scientific spectrophotometer ND-2000, USA). The complementary DNA was synthesized using the thermo scientific RevertAid first Strand cDNA synthesis kit (Fermentas, life sciences USA) according to instructions provided by the kit’s manufacturer. Quantification of the produced cDNA was carried out with the help of nanodrop (Thermo Scientific spectrophotometer ND- 2000, USA) by using 1 µL of DNA sample.
2.3. Amplification of VP4 and VP6 Gene Through PCR
For the confirmation of bovine rotavirus, amplification of both the VP4 and VP6 gene was carried out using primer sets given in Table 1 as mentioned in (4, 16, 17). For the desired amplification of VP4 gene (880 bp) and VP6 gene (250 bp) of bovine rotavirus, optimization of PCR conditions was done by using the varying concentrations of cDNA, primers and Taq polymerase in PCR reaction mixture. Optimized reaction mixture for PCR was cDNA 2.0µL, PCR buffer (2 mM) 2.0 µL, dNTPs (25 mM) 2.5 µL, MgCl2 2.5 µL, VP4/VP6 F (10 pmol) 3.0 µL, VP4/VP6 R (10 pmol) 6.0 µL, Taq Polymerase 0.5 µL and water 6.5 µL. Conditions for the amplification of VP4 gene of bovine rotavirus include initial denaturation at 95°C for 1 minute, followed by denaturation at 94°C for 30 seconds, annealing at 46.3°C for 30 seconds, extension at 72°C for 1 minute and final extension at 72°C for 10 minutes. Similarly, conditions for the amplification of the VP6 gene of bovine rotavirus include initial denaturation at 95°C for 4 minutes, followed by denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and final extension at 72°C for 10 minutes. Steps 2 - 4 were repeated 35 times, to get the required product in both VP4 and VP6 gene amplification. To analyze the PCR amplification, agarose gel electrophoresis was performed. For this, 1.2% gel was prepared and 1μL of 100 bp DNA ladder along with the PCR product was run at 110 volts for 45 minutes. The size of the PCR product for VP4 gene (880 bp) and VP6 gene (250 bp) was illuminated in a gel documentation system and a photograph was taken.
| Primer Name | Sequence, 5’ - 3’ | Melting Temp, °C | Primer Length | Amplicon Size |
|---|---|---|---|---|
| VP4-F | TGGCTTCGCTCATTTATAGACA | 54.2 | 22 | 880 bp |
| VP4-R | ATTTCGGACCATTTATAACC | 47.4 | 20 | |
| VP6-F | GGCTTTAAAACGAAGTCTTC | 50 | 20 | 250 bp |
| VP6-R | GGTCACATCCTCTCACT | 45 | 17 |
Primers for Amplification of the VP4 and VP6 Gene of Bovine Rotavirus
2.4. Sequencing and Bioinformatics Analysis
After precipitation, Sequencing of the PCR products were done using ABI Prism 3100 genetic analyzer (Applied Biosystems, Inc., Foster City, CA, USA) by Sanger Chain termination method. Homology analysis was made with the help of BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analysis was performed to find out evolutionary relationship using the software known as MEGA 5.1 (18).
3. Results
In this study, antigen capture sandwich ELISA of 200 diarrheic fecal samples showed that 12 samples (6%) were positive for the presence of bovine rotavirus. Out of these samples, 7/12 was from buffalo and 5/12 from cattle calves. Amongst these positive samples, 3 cattle and 5 buffalo calves were from the Lahore district while 2 cattle and 2 buffalo calves were from the Faisalabad district.
After RNA extraction and cDNA synthesis, the PCR was done for amplification of VP4 gene and VP6 gene of all ELISA positive bovine rotavirus samples. The PCR resulted in amplification of 880 bp VP4 gene in 5 samples, when visualized on 1.2% agarose gel. Out of these 5 samples, 3 were from buffalo calf and 2 were from cattle calf. The PCR resulted in amplification of 250 bp VP6 gene in 4 samples, when visualized on 1.2% agarose gel. Out of these 4 samples, 3 were from buffalo calf and 1 was from cattle calf.
3.1. Phylogenetic Analysis
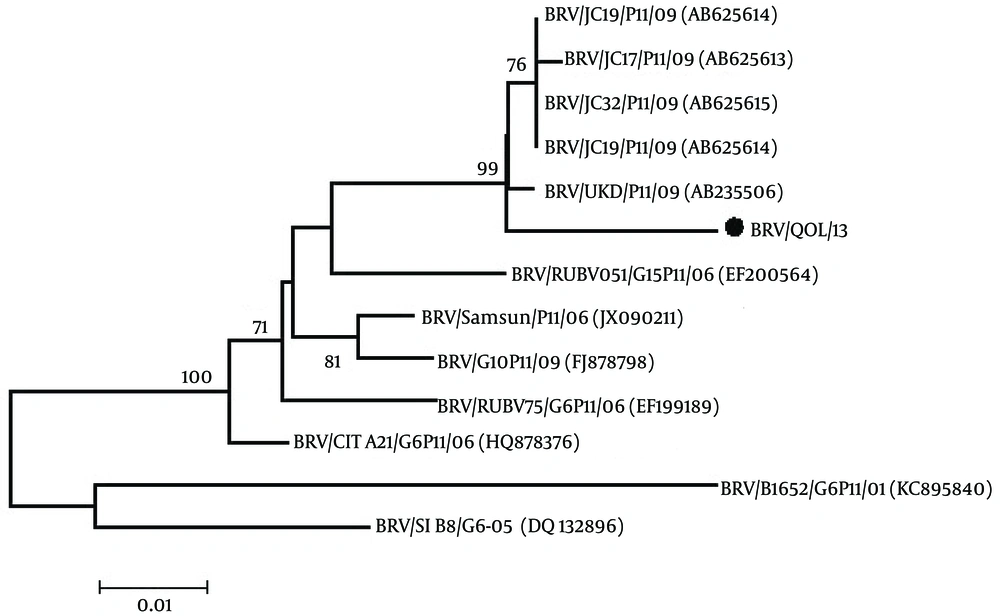
Phylogenetic analysis of the VP4 gene of bovine rotavirus was done with the other 12 VP4 genes of bovine rotavirus reported on NCBI using the MEGA 5.1 software (18). The neighbor-joining tree having the bootstrap values (1000 replicates) was constructed and analyzed. Bootstrap values above 50 were shown.
From the phylogenetic tree in Figure 1, it is evident that the Pakistani bovine rotavirus VP4 gene (BRV/QOL/13; Accession no. KX537745) has a maximum identity of 98% with Indian bovine rotaviruses VP4 gene (accession No. AB625614, AB625613, AB625615, AB625616). A 96% homology with Indian bovine rotaviruses VP4 gene (accession No. EF200564), Turkey bovine rotaviruses VP4 gene (accession No. JX090211) and Ireland bovine rotaviruses VP4 gene (accession No. HQ878376). A 95% homology with Turkey bovine rotaviruses VP4 gene (accession No. FJ878798) and Indian bovine rotaviruses VP4 gene (accession No. EF199489). A 92% homology with Slovenia bovine rotaviruses VP4 gene (accession No. DQ132896) and a 90% homology with Argentina bovine rotaviruses VP4 gene (accession No. KC895840).
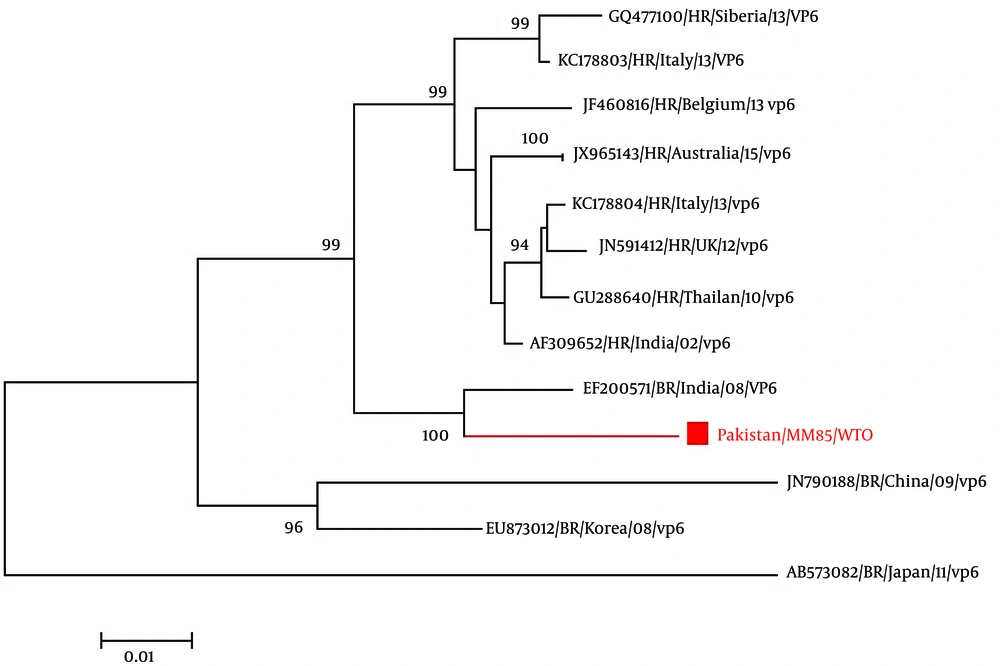
For the phylogenetic analysis of the VP6 gene of bovine rotavirus, inference of the gene’s evolutionary history was done through the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 0.19558277 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (19). The evolutionary distances of the data were computed using the Kimura 2-parameter method (K80) and are in the units of the number of base substitutions per site. A total of 29 nucleotide sequences were involved in the analysis. Codon positions included were 1st + 2nd + 3rd + Noncoding. Elimination of the positions having missing data and gaps was done. There were a total of 1,158 positions in the final dataset. Evolutionary analysis was done through the use of software known as MEGA6 (20).
The phylogenetic tree of VP6 gene of bovine rotavirus (Figure 2) compared with other 28 VP6 gene of rotavirus reported on NCBI. In the phylogenetic tree, rotavirus used for bovine rotavirus comparison are followed by strain name, genotype, year of submission and accession no. From the phylogenetic tree, Figure 2, it is evident that Pakistani bovine rotavirus VP6 gene (Pakistan/ MM85/ QOL; Accession No. KX537746) has a maximum identity of 98% with Indian bovine rotaviruses A VP6 gene (Accession No. JF720873, EF200565) and 96% homology with bovine rotavirus A gene from Indian (Accession No. EF200568 and EF200571).
4. Discussion
Neonatal calf diarrhea is a prime disease affecting newborn calves leading to morbidity and mortality in newborn calves, causing economic losses due to the costs of treatment, diagnostics and poor growth performance (13). A crucial period for these calves is the first few days following birth. In developing countries like Pakistan, domestic animals are the major income source for poor families. These families suffered badly due to the neonatal calf mortality curse. Among numerous viral, bacterial and parasitic causative agents, bovine rotavirus is the foremost cause of neonatal calf diarrhea in domestic animals. The cause of neonatal calf mortality is specifically related to group A bovine rotaviruses (21).
After sequencing and bioinformatics analysis, the phylogenetic tree was constructed through the methods known as neighbor-joining, which was the same as constructed by Abe et al., 2011 (2). By observing the results of the phylogenetic analysis, it is evident that the Pakistani bovine rotavirus VP4 gene (BRV/ QOL/ 13, accession No. KX537745) has a maximum identity of 98% with Indian bovine rotaviruses VP4 gene (accession No. AB625614, AB625613, AB625615, AB625616), which has a basis on the fact that India is a close geographical neighbor. It is important to note that although homology is very high, there are many differences between nucleotides of the Pakistani bovine rotavirus VP4 gene compared to that of the Indian bovine rotavirus VP4 gene. The P type is P11 of these closely homologous Indian bovine rotavirus VP4 genes, so keeping the recorded results of this study in mind can be assumed that our Pakistani bovine rotavirus VP4 gene also has a P11, P type.
Phylogenetic analysis of the VP6 gene of bovine rotavirus (Pakistan/ MM85/ QOL, Accession No. KX537746) revealed that it has maximum of 100% identity with Indian bovine rotaviruses VP6 gene (Accession No. JF720873). The inner capsid protein VP6 has 62%, 94% and 98% similarity with Indian Bovine rotavirus A strain VP6 gene (Accession No. EF200565, Accession No. EF200568 and Accession No. EF200571), respectively. These current findings of VP6 inner capsid gene resembled more with the Indian bovine rotavirus suggesting that being a neighboring country and sharing boundaries of the regions same type and strain of bovine rotavirus is circulating in Pakistan. The group of rotavirus on basis of VP6 gene phylogenetic analysis showed that it belongs to group A and subgroup 1 bovine rotavirus. As known that VP6 is the major structural component of the rotavirus capsid. It also has an important role when it comes to the virion structure. This is because it interacts with the outer capsid proteins known as VP4 and VP7, as well as the core protein known as VP2 (21).
Bovine rotavirus presents 6% (Mukhtar et al., 2017 paper in press) of calf diarrhea in Pakistani cattle and buffalo samples. The VP4 and VP6 genes show closed relationships with bovine rotavirus reported from Indian isolates.